Postmitotic centriole disengagement and maturation leads to centrosome amplification in polyploid trophoblast giant cells
- PMID: 36001376
- PMCID: PMC9634975
- DOI: 10.1091/mbc.E22-05-0182
Postmitotic centriole disengagement and maturation leads to centrosome amplification in polyploid trophoblast giant cells
Abstract
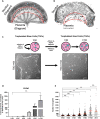
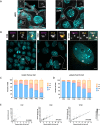
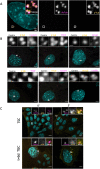
DNA replication is normally coupled with centriole duplication in the cell cycle. Trophoblast giant cells (TGCs) of the placenta undergo endocycles resulting in polyploidy but their centriole state is not known. We used a cell culture model for TGC differentiation to examine centriole and centrosome number and properties. Before differentiation, trophoblast stem cells (TSCs) have either two centrioles before duplication or four centrioles after. We find that the average nuclear area increases approximately eight-fold over differentiation, but most TGCs do not have more than four centrioles. However, these centrioles become disengaged, acquire centrosome proteins, and can nucleate microtubules. In addition, some TGCs undergo further duplication and disengagement of centrioles, resulting in substantially higher numbers. Live imaging revealed that disengagement and separation are centriole autonomous and can occur asynchronously. Centriole amplification, when present, occurs by the standard mechanism of one centriole generating one procentriole. PLK4 inhibition blocks centriole formation in differentiating TGCs but does not affect endocycle progression. In summary, centrioles in TGC endocycles undergo disengagement and conversion to centrosomes. This increases centrosome number but to a limited extent compared with DNA reduplication.
Figures



References
-
- Carney EW, Prideaux V, Lye SJ, Rossant J (1993). Progressive expression of trophoblast-specific genes during formation of mouse trophoblast giant cells in vitro. Mol Reprod Dev 34, 357–368. - PubMed
-
- Carvalho AF, Klisch K, Miglino MA, Pereira FTV, Bevilacqua E (2006). Binucleate trophoblast giant cells in the water buffalo (Bubalus bubalis) placenta. J Morphol 267, 50–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

