Long noncoding RNA CHROMR regulates antiviral immunity in humans
- PMID: 36001732
- PMCID: PMC9477407
- DOI: 10.1073/pnas.2210321119
Long noncoding RNA CHROMR regulates antiviral immunity in humans
Abstract
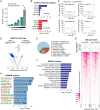
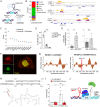
Long noncoding RNAs (lncRNAs) have emerged as critical regulators of gene expression, yet their contribution to immune regulation in humans remains poorly understood. Here, we report that the primate-specific lncRNA CHROMR is induced by influenza A virus and SARS-CoV-2 infection and coordinates the expression of interferon-stimulated genes (ISGs) that execute antiviral responses. CHROMR depletion in human macrophages reduces histone acetylation at regulatory regions of ISG loci and attenuates ISG expression in response to microbial stimuli. Mechanistically, we show that CHROMR sequesters the interferon regulatory factor (IRF)-2-dependent transcriptional corepressor IRF2BP2, thereby licensing IRF-dependent signaling and transcription of the ISG network. Consequently, CHROMR expression is essential to restrict viral infection of macrophages. Our findings identify CHROMR as a key arbitrator of antiviral innate immune signaling in humans.
Keywords: antiviral response; innate immune signaling; interferon-stimulated genes; lncRNA.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

