Optimized human intestinal organoid model reveals interleukin-22-dependency of paneth cell formation
- PMID: 36002022
- PMCID: PMC9438971
- DOI: 10.1016/j.stem.2022.08.002
Optimized human intestinal organoid model reveals interleukin-22-dependency of paneth cell formation
Erratum in
-
Optimized human intestinal organoid model reveals interleukin-22-dependency of paneth cell formation.Cell Stem Cell. 2022 Dec 1;29(12):1718-1720. doi: 10.1016/j.stem.2022.11.001. Cell Stem Cell. 2022. PMID: 36459971 Free PMC article. No abstract available.
Abstract
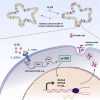
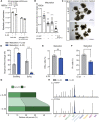
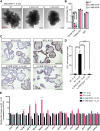
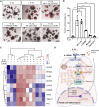
Opposing roles have been proposed for IL-22 in intestinal pathophysiology. We have optimized human small intestinal organoid (hSIO) culturing, constitutively generating all differentiated cell types while maintaining an active stem cell compartment. IL-22 does not promote the expansion of stem cells but rather slows the growth of hSIOs. In hSIOs, IL-22 is required for formation of Paneth cells, the prime producers of intestinal antimicrobial peptides (AMPs). Introduction of inflammatory bowel disease (IBD)-associated loss-of-function mutations in the IL-22 co-receptor gene IL10RB resulted in abolishment of Paneth cells in hSIOs. Moreover, IL-22 induced expression of host defense genes (such as REG1A, REG1B, and DMBT1) in enterocytes, goblet cells, Paneth cells, Tuft cells, and even stem cells. Thus, IL-22 does not directly control the regenerative capacity of crypt stem cells but rather boosts Paneth cell numbers, as well as the expression of AMPs in all cell types.
Keywords: IL-22; IL10RB; Paneth cells; anti-microbial proteins; enterocytes; inflammatory bowel disease; intestinal stem cells; mTOR; organoids; regeneration.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests H.C. is an inventor of several patents related to organoid technology; his full disclosure is given at https://www.uu.nl/staff/JCClevers/.
Figures



References
-
- Angerer P., Haghverdi L., Büttner M., Theis F.J., Marr C., Buettner F. Destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. - PubMed
-
- Artegiani B., Hendriks D., Beumer J., Kok R., Zheng X., Joore I., Chuva de Sousa Lopes S., van Zon J., Tans S., Clevers H. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat. Cell Biol. 2020;22:321–331. - PubMed
-
- Begue B., Verdier J., Rieux-Laucat F., Goulet O., Morali A., Canioni D., Hugot J.P., Daussy C., Verkarre V., Pigneur B., et al. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am. J. Gastroenterol. 2011;106:1544–1555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

