Snail-regulated exosomal microRNA-21 suppresses NLRP3 inflammasome activity to enhance cisplatin resistance
- PMID: 36002186
- PMCID: PMC9413180
- DOI: 10.1136/jitc-2022-004832
Snail-regulated exosomal microRNA-21 suppresses NLRP3 inflammasome activity to enhance cisplatin resistance
Abstract
Background: Compared with the precise targeting of drug-resistant mutant cancer cells, strategies for eliminating non-genetic adaptation-mediated resistance are limited. The pros and cons of the existence of inflammasomes in cancer have been reported. Nevertheless, the dynamic response of inflammasomes to therapies should be addressed.
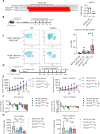
Methods: Tumor-derived exosomes were purified by differential ultracentrifugation and validated by nanoparticle tracking analysis and transmission electron microscopy. A proximity ligation assay and interleukin-1β (IL-1β) level were used for detecting activation of NLRP3 inflammasomes. RNA sequencing was used to analyze the exosomal RNAs. MIR21 knocked out human monocytic THP cells and mir21 knocked out murine oral cancer MTCQ1 cells were generated for confirming the exosomal delivery of microRNA (miR)-21. Syngeneic murine models for head and neck cancer (C57BLJ/6J), breast cancer (BALB/C) and lung cancer (C57BL/6J) were applied for examining the impact of Snail-miR21 axis on inflammasome activation in vivo. Single-cell RNA sequencing was used for analyzing the tumor-infiltrated immune cells. Head and neck patient samples were used for validating the findings in clinical samples.
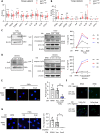
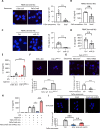
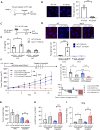
Results: We demonstrated that in cancer cells undergoing Snail-induced epithelial-mesenchymal transition (EMT), tumor cells suppress NLRP3 inflammasome activities of tumor-associated macrophages (TAMs) in response to chemotherapy through the delivery of exosomal miR-21. Mechanistically, miR-21 represses PTEN and BRCC3 to facilitate NLRP3 phosphorylation and lysine-63 ubiquitination, inhibiting NLRP3 inflammasome assembly. Furthermore, the Snail-miR-21 axis shapes the post-chemotherapy tumor microenvironment (TME) by repopulating TAMs and by activating CD8+ T cells. In patients with head and neck cancer, the Snail-high cases lacked post-chemotherapy IL-1β surge and were correlated with a worse response.
Conclusions: This finding reveals the mechanism of EMT-mediated resistance beyond cancer stemness through modulation of post-treatment inflammasome activity. It also highlights the dynamic remodeling of the TME throughout metastatic evolution.
Keywords: head and neck neoplasms; immunity, innate; macrophages; tumor microenvironment.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
