Identification and construction of a multi-epitopes vaccine design against Klebsiella aerogenes: molecular modeling study
- PMID: 36002561
- PMCID: PMC9399595
- DOI: 10.1038/s41598-022-18610-0
Identification and construction of a multi-epitopes vaccine design against Klebsiella aerogenes: molecular modeling study
Abstract
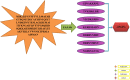
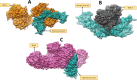
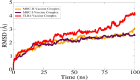
A rapid rise in antibiotic resistance by bacterial pathogens is due to these pathogens adaptation to the changing environmental conditions. Antibiotic resistance infections can be reduced by a number of ways such as development of safe and effective vaccine. Klebsiella aerogene is a gram-negative, rod-shaped bacterium resistant to a variety of antibiotics and no commercial vaccine is available against the pathogen. Identifying antigens that can be easily evaluated experimentally would be crucial to successfully vaccine development. Reverse vaccinology (RV) was used to identify vaccine candidates based on complete pathogen proteomic information. The fully sequenced proteomes include 44,115 total proteins of which 43,316 are redundant and 799 are non-redundant. Subcellular localization showed that only 1 protein in extracellular matrix, 7 were found in outer-membrane proteins, and 27 in the periplasm space. A total of 3 proteins were found virulent. Next in the B-cell-derived T-cell epitopes mapping phase, the 3 proteins (Fe2+- enterobactin, ABC transporter substrate-binding protein, and fimbriae biogenesis outer membrane usher protein) were tested positive for antigenicity, toxicity, and solubility. GPGPG linkers were used to prepare a vaccine construct composed of 7 epitopes and an adjuvant of toxin B subunit (CTBS). Molecular docking of vaccine construct with major histocompatibility-I (MHC-I), major histocompatibility-II (MHC-II), and Toll-like receptor 4 (TLR4) revealed vaccine robust interactions and stable binding pose to the receptors. By using molecular dynamics simulations, the vaccine-receptors complexes unveiled stable dynamics and uniform root mean square deviation (rmsd). Further, binding energies of complex were computed that again depicted strong intermolecular bindings and formation of stable conformation.
© 2022. The Author(s).
Conflict of interest statement
The author declares no competing interests.
Figures





Similar articles
-
Reverse Vaccinology and Immunoinformatic Assisted Designing of a Multi-Epitopes Based Vaccine Against Nosocomial Burkholderia cepacia.Front Microbiol. 2022 Jun 28;13:929400. doi: 10.3389/fmicb.2022.929400. eCollection 2022. Front Microbiol. 2022. PMID: 35875518 Free PMC article.
-
A computational quest for identifying potential vaccine candidates against Moraxella lacunata: a multi-pronged approach.J Biomol Struct Dyn. 2024 Apr;42(6):2976-2989. doi: 10.1080/07391102.2023.2212793. Epub 2023 May 12. J Biomol Struct Dyn. 2024. PMID: 37177816
-
Computational Design of a Chimeric Vaccine against Plesiomonas shigelloides Using Pan-Genome and Reverse Vaccinology.Vaccines (Basel). 2022 Nov 8;10(11):1886. doi: 10.3390/vaccines10111886. Vaccines (Basel). 2022. PMID: 36366394 Free PMC article.
-
Computational exploration and design of a multi-epitopes vaccine construct against Chlamydia psittaci.J Biomol Struct Dyn. 2024;42(22):12105-12121. doi: 10.1080/07391102.2023.2268173. Epub 2023 Oct 28. J Biomol Struct Dyn. 2024. PMID: 37897717
-
Harnessing immunopeptidomics for next-generation vaccines against intracellular bacterial pathogens.Mol Biol Rep. 2025 Jun 7;52(1):561. doi: 10.1007/s11033-025-10678-x. Mol Biol Rep. 2025. PMID: 40481904 Review. No abstract available.
Cited by
-
Design of a Multi-Epitope Vaccine against Tuberculosis from Mycobacterium tuberculosis PE_PGRS49 and PE_PGRS56 Proteins by Reverse Vaccinology.Microorganisms. 2023 Jun 24;11(7):1647. doi: 10.3390/microorganisms11071647. Microorganisms. 2023. PMID: 37512820 Free PMC article.
-
The Promising Potential of Reverse Vaccinology-Based Next-Generation Vaccine Development over Conventional Vaccines against Antibiotic-Resistant Bacteria.Vaccines (Basel). 2023 Jul 20;11(7):1264. doi: 10.3390/vaccines11071264. Vaccines (Basel). 2023. PMID: 37515079 Free PMC article. Review.
-
Proteomics-based vaccine targets annotation and design of multi-epitope vaccine against antibiotic-resistant Streptococcus gallolyticus.Sci Rep. 2024 Feb 28;14(1):4836. doi: 10.1038/s41598-024-55372-3. Sci Rep. 2024. PMID: 38418560 Free PMC article.
-
An immunoinformatics and structural vaccinology approach to design a novel and potent multi-epitope base vaccine targeting Zika virus.BMC Chem. 2024 Feb 13;18(1):31. doi: 10.1186/s13065-024-01132-3. BMC Chem. 2024. PMID: 38350946 Free PMC article.
-
Core Proteomics and Immunoinformatic Approaches to Design a Multiepitope Reverse Vaccine Candidate against Chagas Disease.Vaccines (Basel). 2022 Oct 7;10(10):1669. doi: 10.3390/vaccines10101669. Vaccines (Basel). 2022. PMID: 36298534 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

