Natural switches in behaviour rapidly modulate hippocampal coding
- PMID: 36002570
- PMCID: PMC9433324
- DOI: 10.1038/s41586-022-05112-2
Natural switches in behaviour rapidly modulate hippocampal coding
Abstract
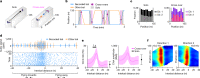
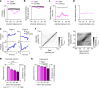
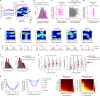
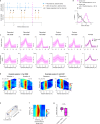
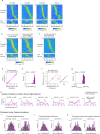
Throughout their daily lives, animals and humans often switch between different behaviours. However, neuroscience research typically studies the brain while the animal is performing one behavioural task at a time, and little is known about how brain circuits represent switches between different behaviours. Here we tested this question using an ethological setting: two bats flew together in a long 135 m tunnel, and switched between navigation when flying alone (solo) and collision avoidance as they flew past each other (cross-over). Bats increased their echolocation click rate before each cross-over, indicating attention to the other bat1-9. Hippocampal CA1 neurons represented the bat's own position when flying alone (place coding10-14). Notably, during cross-overs, neurons switched rapidly to jointly represent the interbat distance by self-position. This neuronal switch was very fast-as fast as 100 ms-which could be revealed owing to the very rapid natural behavioural switch. The neuronal switch correlated with the attention signal, as indexed by echolocation. Interestingly, the different place fields of the same neuron often exhibited very different tuning to interbat distance, creating a complex non-separable coding of position by distance. Theoretical analysis showed that this complex representation yields more efficient coding. Overall, our results suggest that during dynamic natural behaviour, hippocampal neurons can rapidly switch their core computation to represent the relevant behavioural variables, supporting behavioural flexibility.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures















References
-
- Simmons JA, Fenton MB, O’Farrell MJ. Echolocation and pursuit of prey by bats. Science. 1979;203:16–21. - PubMed
-
- Petrites AE, Eng OS, Mowlds DS, Simmons JA, DeLong CM. Interpulse interval modulation by echolocating big brown bats (Eptesicus fuscus) in different densities of obstacle clutter. J. Comp. Physiol. A. 2009;195:603–617. - PubMed
-
- Yovel Y, Falk B, Moss CF, Ulanovsky N. Optimal localization by pointing off axis. Science. 2010;327:701–704. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

