Phosphorylation of muramyl peptides by NAGK is required for NOD2 activation
- PMID: 36002575
- PMCID: PMC9477735
- DOI: 10.1038/s41586-022-05125-x
Phosphorylation of muramyl peptides by NAGK is required for NOD2 activation
Abstract
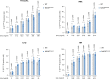
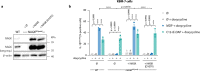
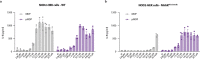
Bacterial cell wall components provide various unique molecular structures that are detected by pattern recognition receptors (PRRs) of the innate immune system as non-self. Most bacterial species form a cell wall that consists of peptidoglycan (PGN), a polymeric structure comprising alternating amino sugars that form strands cross-linked by short peptides. Muramyl dipeptide (MDP) has been well documented as a minimal immunogenic component of peptidoglycan1-3. MDP is sensed by the cytosolic nucleotide-binding oligomerization domain-containing protein 24 (NOD2). Upon engagement, it triggers pro-inflammatory gene expression, and this functionality is of critical importance in maintaining a healthy intestinal barrier function5. Here, using a forward genetic screen to identify factors required for MDP detection, we identified N-acetylglucosamine kinase (NAGK) as being essential for the immunostimulatory activity of MDP. NAGK is broadly expressed in immune cells and has previously been described to contribute to the hexosamine biosynthetic salvage pathway6. Mechanistically, NAGK functions upstream of NOD2 by directly phosphorylating the N-acetylmuramic acid moiety of MDP at the hydroxyl group of its C6 position, yielding 6-O-phospho-MDP. NAGK-phosphorylated MDP-but not unmodified MDP-constitutes an agonist for NOD2. Macrophages from mice deficient in NAGK are completely deficient in MDP sensing. These results reveal a link between amino sugar metabolism and innate immunity to bacterial cell walls.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











References
-
- Kotani S, Watanabe Y, Kinoshita F, Shimono T, Morisaki I. Immunoadjuvant activities of synthetic N-acetyl-muramyl-peptides or -amino acids. Biken J. 1975;18:105–111. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

