Biophysical, Molecular and Proteomic Profiling of Human Retinal Organoid-Derived Exosomes
- PMID: 36002615
- PMCID: PMC10576571
- DOI: 10.1007/s11095-022-03350-7
Biophysical, Molecular and Proteomic Profiling of Human Retinal Organoid-Derived Exosomes
Abstract
Purpose: There is a growing interest in extracellular vesicles (EVs) for ocular applications as therapeutics, biomarkers, and drug delivery vehicles. EVs secreted from mesenchymal stem cells (MSCs) have shown to provide therapeutic benefits in ocular conditions. However, very little is known about the properties of bioreactor cultured-3D human retinal organoids secreted EVs. This study provides a comprehensive morphological, nanomechanical, molecular, and proteomic characterization of retinal organoid EVs and compares it with human umbilical cord (hUC) MSCs.
Methods: The morphology and nanomechanical properties of retinal organoid EVs were assessed using Nanoparticle tracking analysis (NTA) and Atomic force microscopy (AFM). Gene expression analysis of exosome biogenesis of early and late retinal organoids were compared using qPCR. The protein profile of the EVs were analyzed with proteomic tools.
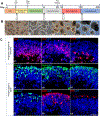
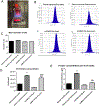
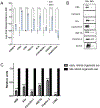
Results: NTA indicated the average size of EV as 100-250 nm. A high expression of exosome biogenesis genes was observed in late retinal organoids EVs. Immunoblot analysis showed highly expressed exosomal markers in late retinal organoids EVs compared to early retinal organoids EVs. Protein profiling of retinal organoid EVs displayed a higher differential expression of retinal function-related proteins and EV biogenesis proteins than hUCMSC EVs, implicating that the use of retinal organoid EVs may have a superior therapeutic effect on retinal disorders.
Conclusion: This study provides supplementary knowledge on the properties of retinal organoid EVs and suggests their potential use in the diagnostic and therapeutic treatments for ocular diseases.
Keywords: Bioreactor; Extracellular vesicles; Human retinal organoids; Proteomics; Stem cells.
© 2022. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Figures


References
-
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. The Lancet. 2006;368(9549):1795–809. - PubMed
-
- Goodwin P. Hereditary retinal disease. Curr Opin Ophthalmol. 2008;19(3):255–62. - PubMed
-
- Hamel J-F. Les ruines du progrès chez Walter Benjamin: anticipation futuriste, fausse reconnaissance et politique du présent. Protée. 2007;35(2):7–14.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

