Analysis of KIR gene variants in The Cancer Genome Atlas and UK Biobank using KIRCLE
- PMID: 36002830
- PMCID: PMC9400285
- DOI: 10.1186/s12915-022-01392-2
Analysis of KIR gene variants in The Cancer Genome Atlas and UK Biobank using KIRCLE
Abstract
Background: Natural killer (NK) cells represent a critical component of the innate immune system's response against cancer and viral infections, among other diseases. To distinguish healthy host cells from infected or tumor cells, killer immunoglobulin receptors (KIR) on NK cells bind and recognize Human Leukocyte Antigen (HLA) complexes on their target cells. However, NK cells exhibit great diversity in their mechanism of activation, and the outcomes of their activation are not yet understood fully. Just like the HLAs they bind, KIR receptors exhibit high allelic diversity in the human population. Here we provide a method to identify KIR allele variants from whole exome sequencing data and uncover novel associations between these variants and various molecular and clinical correlates.
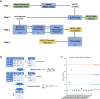
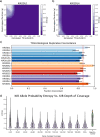
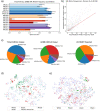
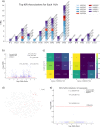
Results: In order to better understand KIRs, we have developed KIRCLE, a novel method for genotyping individual KIR genes from whole exome sequencing data, and used it to analyze approximately sixty-thousand patient samples in The Cancer Genome Atlas (TCGA) and UK Biobank. We were able to assess population frequencies for different KIR alleles and demonstrate that, similar to HLA alleles, individuals' KIR alleles correlate strongly with their ethnicities. In addition, we observed associations between different KIR alleles and HLA alleles, including HLA-B*53 with KIR3DL2*013 (Fisher's exact FDR = 7.64e-51). Finally, we showcased statistically significant associations between KIR alleles and various clinical correlates, including peptic ulcer disease (Fisher's exact FDR = 0.0429) and age of onset of atopy (Mann-Whitney U FDR = 0.0751).
Conclusions: We show that KIRCLE is able to infer KIR variants accurately and consistently, and we demonstrate its utility using data from approximately sixty-thousand individuals from TCGA and UK Biobank to discover novel molecular and clinical correlations with KIR germline variants. Peptic ulcer disease and atopy are just two diseases in which NK cells may play a role beyond their "classical" realm of anti-tumor and anti-viral responses. This tool may be used both as a benchmark for future KIR-variant-inference algorithms, and to better understand the immunogenomics of and disease processes involving KIRs.
Keywords: Immunogenomics; Killer immunoglobulin receptors; Natural killer cells.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Allelic Polymorphisms of KIRs and HLAs Predict Favorable Responses to Tyrosine Kinase Inhibitors in CML.Cancer Immunol Res. 2018 Jun;6(6):745-754. doi: 10.1158/2326-6066.CIR-17-0462. Epub 2018 Apr 25. Cancer Immunol Res. 2018. PMID: 29695383
-
The Impact of Killer Cell Immunoglobulin-Like Receptors and Human Leukocyte Antigen-E, Human Leukocyte Antigen-G Polymorphisms on Innate Immunity and COVID-19 Severity.J Immunol Res. 2025 May 12;2025:6691437. doi: 10.1155/jimr/6691437. eCollection 2025. J Immunol Res. 2025. PMID: 40391199 Free PMC article.
-
Next-generation sequencing technology a new tool for killer cell immunoglobulin-like receptor allele typing in hematopoietic stem cell transplantation.Transfus Clin Biol. 2018 Feb;25(1):87-89. doi: 10.1016/j.tracli.2017.07.005. Epub 2017 Oct 12. Transfus Clin Biol. 2018. PMID: 29032017
-
[Role of combination NK/KIRs in the natural history of viral infections.].Recenti Prog Med. 2017 Jul-Aug;108(7):333-337. doi: 10.1701/2731.27839. Recenti Prog Med. 2017. PMID: 28845855 Review. Italian.
-
KIR genes and their role in spondyloarthropathies.Adv Exp Med Biol. 2009;649:286-99. doi: 10.1007/978-1-4419-0298-6_22. Adv Exp Med Biol. 2009. PMID: 19731638 Review.
Cited by
-
kir-mapper: A Toolkit for Killer-Cell Immunoglobulin-Like Receptor (KIR) Genotyping From Short-Read Second-Generation Sequencing Data.HLA. 2025 Mar;105(3):e70092. doi: 10.1111/tan.70092. HLA. 2025. PMID: 40095784
-
Full-resolution HLA and KIR gene annotations for human genome assemblies.Genome Res. 2024 Nov 20;34(11):1931-1941. doi: 10.1101/gr.278985.124. Genome Res. 2024. PMID: 38839374 Free PMC article.
-
Exploration of KIR genes and hematological-related diseases in Chinese Han population.Sci Rep. 2023 Jun 16;13(1):9773. doi: 10.1038/s41598-023-36882-y. Sci Rep. 2023. PMID: 37328612 Free PMC article.
-
Full resolution HLA and KIR genes annotation for human genome assemblies.bioRxiv [Preprint]. 2024 Jan 23:2024.01.20.576452. doi: 10.1101/2024.01.20.576452. bioRxiv. 2024. Update in: Genome Res. 2024 Nov 20;34(11):1931-1941. doi: 10.1101/gr.278985.124. PMID: 38328160 Free PMC article. Updated. Preprint.
-
Innate receptors modulating adaptive T cell responses: KIR-HLA interactions and T cell-mediated control of chronic viral infections.Immunogenetics. 2023 Jun;75(3):269-282. doi: 10.1007/s00251-023-01293-w. Epub 2023 Jan 31. Immunogenetics. 2023. PMID: 36719466 Free PMC article. Review.
References
-
- Kamoda Y, Uematsu H, Yoshihara A, Miyazaki H, Senpuku H. Role of activated natural killer cells in oral diseases. Jpn J Infect Dis. 2008;61(6):469–474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

