Charge-based interactions through peptide position 4 drive diversity of antigen presentation by human leukocyte antigen class I molecules
- PMID: 36003074
- PMCID: PMC9391200
- DOI: 10.1093/pnasnexus/pgac124
Charge-based interactions through peptide position 4 drive diversity of antigen presentation by human leukocyte antigen class I molecules
Abstract
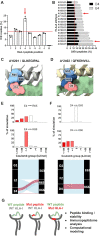
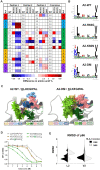
Human leukocyte antigen class I (HLA-I) molecules bind and present peptides at the cell surface to facilitate the induction of appropriate CD8+ T cell-mediated immune responses to pathogen- and self-derived proteins. The HLA-I peptide-binding cleft contains dominant anchor sites in the B and F pockets that interact primarily with amino acids at peptide position 2 and the C-terminus, respectively. Nonpocket peptide-HLA interactions also contribute to peptide binding and stability, but these secondary interactions are thought to be unique to individual HLA allotypes or to specific peptide antigens. Here, we show that two positively charged residues located near the top of peptide-binding cleft facilitate interactions with negatively charged residues at position 4 of presented peptides, which occur at elevated frequencies across most HLA-I allotypes. Loss of these interactions was shown to impair HLA-I/peptide binding and complex stability, as demonstrated by both in vitro and in silico experiments. Furthermore, mutation of these Arginine-65 (R65) and/or Lysine-66 (K66) residues in HLA-A*02:01 and A*24:02 significantly reduced HLA-I cell surface expression while also reducing the diversity of the presented peptide repertoire by up to 5-fold. The impact of the R65 mutation demonstrates that nonpocket HLA-I/peptide interactions can constitute anchor motifs that exert an unexpectedly broad influence on HLA-I-mediated antigen presentation. These findings provide fundamental insights into peptide antigen binding that could broadly inform epitope discovery in the context of viral vaccine development and cancer immunotherapy.
Keywords: computational modeling; human leukocyte antigen (HLA); mass spectrometry; molecular dynamics; peptide antigen presentation.
© The Author(s) 2022. Published by Oxford University Press on behalf of the National Academy of Sciences.
Figures



Similar articles
-
HLA-DM catalytically enhances peptide dissociation by sensing peptide-MHC class II interactions throughout the peptide-binding cleft.J Biol Chem. 2020 Mar 6;295(10):2959-2973. doi: 10.1074/jbc.RA119.010645. Epub 2020 Jan 22. J Biol Chem. 2020. PMID: 31969393 Free PMC article.
-
Micropolymorphism outside the peptide-binding groove of human leukocyte antigen (HLA)-C*14 modulates structural stability and shapes immune responses.Int J Biol Macromol. 2025 May;309(Pt 1):142772. doi: 10.1016/j.ijbiomac.2025.142772. Epub 2025 Apr 2. Int J Biol Macromol. 2025. PMID: 40185448
-
Detailed analysis of the effects of Glu/Lys beta69 human leukocyte antigen-DP polymorphism on peptide-binding specificity.Tissue Antigens. 2003 Dec;62(6):459-71. doi: 10.1046/j.1399-0039.2003.00131.x. Tissue Antigens. 2003. PMID: 14617029
-
The pockets guide to HLA class I molecules.Biochem Soc Trans. 2021 Nov 1;49(5):2319-2331. doi: 10.1042/BST20210410. Biochem Soc Trans. 2021. PMID: 34581761 Free PMC article. Review.
-
A comprehensive review and performance evaluation of bioinformatics tools for HLA class I peptide-binding prediction.Brief Bioinform. 2020 Jul 15;21(4):1119-1135. doi: 10.1093/bib/bbz051. Brief Bioinform. 2020. PMID: 31204427 Free PMC article. Review.
Cited by
-
Mutations in glioblastoma proteins do not disrupt epitope presentation and recognition, maintaining a specific CD8 T cell immune response potential.Sci Rep. 2024 Jul 19;14(1):16721. doi: 10.1038/s41598-024-67099-2. Sci Rep. 2024. PMID: 39030304 Free PMC article.
-
APE-Gen2.0: Expanding Rapid Class I Peptide-Major Histocompatibility Complex Modeling to Post-Translational Modifications and Noncanonical Peptide Geometries.J Chem Inf Model. 2024 Mar 11;64(5):1730-1750. doi: 10.1021/acs.jcim.3c01667. Epub 2024 Feb 28. J Chem Inf Model. 2024. PMID: 38415656 Free PMC article.
-
Transfer learning improves pMHC kinetic stability and immunogenicity predictions.Immunoinformatics (Amst). 2024 Mar;13:100030. doi: 10.1016/j.immuno.2023.100030. Epub 2023 Dec 21. Immunoinformatics (Amst). 2024. PMID: 38577265 Free PMC article.
-
Evolution by innovation as a driving force to improve TCR-T therapies.Front Oncol. 2023 Sep 21;13:1216829. doi: 10.3389/fonc.2023.1216829. eCollection 2023. Front Oncol. 2023. PMID: 37810959 Free PMC article. Review.
References
-
- Yewdell JW, Reits E, Neefjes J. 2003. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immunol. 3:952–961. - PubMed
-
- Zinkernagel RM, Doherty PC. 1974. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 248:701–702. - PubMed
-
- Goulmy E, Termijtelen A, Bradley BA, van Rood JJ. 1977. Y-antigen killing by t cells of women is restricted by HLA. Nature. 266:544–545. - PubMed
-
- Kubo RT et al. 1994. Definition of specific peptide motifs for four major HLA-A alleles. J Immunol. 152:3913–3924. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

