Time-series transcriptome analysis identified differentially expressed genes in broiler chicken infected with mixed Eimeria species
- PMID: 36003329
- PMCID: PMC9393255
- DOI: 10.3389/fgene.2022.886781
Time-series transcriptome analysis identified differentially expressed genes in broiler chicken infected with mixed Eimeria species
Abstract
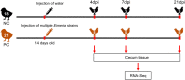
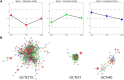
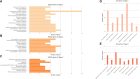
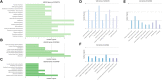
Coccidiosis caused by the Eimeria species is a highly problematic disease in the chicken industry. Here, we used RNA sequencing to observe the time-dependent host responses of Eimeria-infected chickens to examine the genes and biological functions associated with immunity to the parasite. Transcriptome analysis was performed at three time points: 4, 7, and 21 days post-infection (dpi). Based on the changes in gene expression patterns, we defined three groups of genes that showed differential expression. This enabled us to capture evidence of endoplasmic reticulum stress at the initial stage of Eimeria infection. Furthermore, we found that innate immune responses against the parasite were activated at the first exposure; they then showed gradual normalization. Although the cytokine-cytokine receptor interaction pathway was significantly operative at 4 dpi, its downregulation led to an anti-inflammatory effect. Additionally, the construction of gene co-expression networks enabled identification of immunoregulation hub genes and critical pattern recognition receptors after Eimeria infection. Our results provide a detailed understanding of the host-pathogen interaction between chicken and Eimeria. The clusters of genes defined in this study can be utilized to improve chickens for coccidiosis control.
Keywords: Eimeria; anti-inflammation; chicken; gene co-expression network; host-pathogen interaction; innate immunity; transcriptome analysis.
Copyright © 2022 Kim, Chung, Manjula, Seo, Cho, Cho, Ediriweera, Yu, Nam and Lee.
Conflict of interest statement
Author DS was employed by the company Research Institute TNT Research Company. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Major histocompatibility complex genes exhibit a potential immunological role in mixed Eimeria-infected broiler cecum analyzed using RNA sequencing.Anim Biosci. 2024 Jun;37(6):993-1000. doi: 10.5713/ab.23.0412. Epub 2024 Jan 20. Anim Biosci. 2024. PMID: 38271966 Free PMC article.
-
Eimeria maxima-induced transcriptional changes in the cecal mucosa of broiler chickens.Parasit Vectors. 2019 Jun 4;12(1):285. doi: 10.1186/s13071-019-3534-4. Parasit Vectors. 2019. PMID: 31164143 Free PMC article.
-
MicroRNA expression profile of chicken jejunum in different time points Eimeria maxima infection.Front Immunol. 2024 Jan 15;14:1331532. doi: 10.3389/fimmu.2023.1331532. eCollection 2023. Front Immunol. 2024. PMID: 38288128 Free PMC article.
-
Intestinal immune responses to coccidiosis.Dev Comp Immunol. 2000 Mar-Apr;24(2-3):303-24. doi: 10.1016/s0145-305x(99)00080-4. Dev Comp Immunol. 2000. PMID: 10717295 Review.
-
Transgenic Eimeria parasite: A potential control strategy for chicken coccidiosis.Acta Trop. 2020 May;205:105417. doi: 10.1016/j.actatropica.2020.105417. Epub 2020 Feb 24. Acta Trop. 2020. PMID: 32105666 Review.
Cited by
-
Major histocompatibility complex genes exhibit a potential immunological role in mixed Eimeria-infected broiler cecum analyzed using RNA sequencing.Anim Biosci. 2024 Jun;37(6):993-1000. doi: 10.5713/ab.23.0412. Epub 2024 Jan 20. Anim Biosci. 2024. PMID: 38271966 Free PMC article.
-
Transcriptomic analysis of LMH cells in response to the overexpression of a protein of Eimeria tenella encoded by the locus ETH_00028350.Front Vet Sci. 2022 Nov 21;9:1053701. doi: 10.3389/fvets.2022.1053701. eCollection 2022. Front Vet Sci. 2022. PMID: 36478946 Free PMC article.
-
Berberine Reveals Anticoccidial Activity by Influencing Immune Responses in Eimeria acervulina-Infected Chickens.Biomolecules. 2025 Jul 10;15(7):985. doi: 10.3390/biom15070985. Biomolecules. 2025. PMID: 40723856 Free PMC article.
-
Down-regulation of colon mucin production induced by Eimeria pragensis infection in mice.Front Parasitol. 2025 Jun 24;4:1621486. doi: 10.3389/fpara.2025.1621486. eCollection 2025. Front Parasitol. 2025. PMID: 40630939 Free PMC article.
References
LinkOut - more resources
Full Text Sources

