Functional Evaluation and Genetic Landscape of Children and Young Adults Referred for Assessment of Bronchiectasis
- PMID: 36003331
- PMCID: PMC9393783
- DOI: 10.3389/fgene.2022.933381
Functional Evaluation and Genetic Landscape of Children and Young Adults Referred for Assessment of Bronchiectasis
Abstract
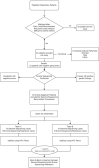
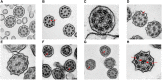
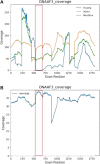
Bronchiectasis is the abnormal dilation of the airway which may be caused by various etiologies in children. Beyond the more recognized cause of bacterial and viral infections and primary immunodeficiencies, other genetic conditions such as cystic fibrosis and primary ciliary dyskinesia (PCD) can also contribute to the disease. Currently, there is still debate on whether genome sequencing (GS) or exome sequencing reanalysis (rES) would be beneficial if the initial targeted testing results returned negative. This study aims to provide a back-to-back comparison between rES and GS to explore the best integrated approach for the functional and genetics evaluation for patients referred for assessment of bronchiectasis. In phase 1, an initial 60 patients were analyzed by exome sequencing (ES) with one additional individual recruited later as an affected sibling for ES. Functional evaluation of the nasal nitric oxide test, transmission electron microscopy, and high-speed video microscopy were also conducted when possible. In phase 2, GS was performed on 30 selected cases with trio samples available. To provide a back-to-back comparison, two teams of genome analysts were alternatively allocated to GS or rES and were blinded to each other's analysis. The time for bioinformatics, analysis, and diagnostic utility was recorded for evaluation. ES revealed five positive diagnoses (5/60, 8.3%) in phase 1, and four additional diagnoses were made by rES and GS (4/30, 13%) during phase 2. Subsequently, one additional positive diagnosis was identified in a sibling by ES and an overall diagnostic yield of 10/61 (16.4%) was reached. Among those patients with a clinical suspicion of PCD (n = 31/61), the diagnostic yield was 26% (n = 8/31). While GS did not increase the diagnostic yield, we showed that a variant of uncertain significance could only be detected by GS due to improved coverage over ES and hence is a potential benefit for GS in the future. We show that genetic testing is an essential component for the diagnosis of early-onset bronchiectasis and is most effective when used in combination with functional tools such as TEM or HSVM. Our comparison of rES vs. GS suggests that rES and GS are comparable in clinical diagnosis.
Keywords: early-onset bronchiectasis; exome sequencing; genome sequencing; high-speed video microscopy; primary ciliary dyskinesia; transmission electron microscopy.
Copyright © 2022 Chau, Lee, Chui, Yu, Fung, Mak, Chau, Siu, Hung, Yeung, Kwong, O'Callaghan, Lau, Lee, Chung and Lee.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Cost-effectiveness analysis of three algorithms for diagnosing primary ciliary dyskinesia: a simulation study.Orphanet J Rare Dis. 2019 Jun 13;14(1):142. doi: 10.1186/s13023-019-1116-3. Orphanet J Rare Dis. 2019. PMID: 31196140 Free PMC article.
-
Diagnostic Yield of Genome Sequencing Versus Exome Sequencing in Pediatric Patients With Rare Phenotypes: A Systematic Review and Meta-Analysis.Am J Med Genet A. 2025 Jun 16:e64146. doi: 10.1002/ajmg.a.64146. Online ahead of print. Am J Med Genet A. 2025. PMID: 40519120 Review.
-
Primary ciliary dyskinesia: From diagnosis to molecular mechanisms.J Pediatr Genet. 2014 Jun;3(2):115-27. doi: 10.3233/PGE-14088. J Pediatr Genet. 2014. PMID: 27625868 Free PMC article.
-
Cost-effectiveness of exome and genome sequencing for children with rare and undiagnosed conditions.Genet Med. 2022 Jun;24(6):1349-1361. doi: 10.1016/j.gim.2022.03.005. Epub 2022 Apr 8. Genet Med. 2022. PMID: 35396982
-
Meta-analysis of the diagnostic and clinical utility of exome and genome sequencing in pediatric and adult patients with rare diseases across diverse populations.Genet Med. 2023 Sep;25(9):100896. doi: 10.1016/j.gim.2023.100896. Epub 2023 May 13. Genet Med. 2023. PMID: 37191093 Review.
Cited by
-
Prenatal genetic diagnosis of fetuses with dextrocardia using whole exome sequencing in a tertiary center.Sci Rep. 2024 Jul 15;14(1):16266. doi: 10.1038/s41598-024-67164-w. Sci Rep. 2024. PMID: 39009665 Free PMC article.
References
LinkOut - more resources
Full Text Sources

