Roles and functions of SARS-CoV-2 proteins in host immune evasion
- PMID: 36003396
- PMCID: PMC9394213
- DOI: 10.3389/fimmu.2022.940756
Roles and functions of SARS-CoV-2 proteins in host immune evasion
Abstract
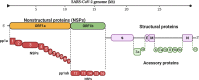
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) evades the host immune system through a variety of regulatory mechanisms. The genome of SARS-CoV-2 encodes 16 non-structural proteins (NSPs), four structural proteins, and nine accessory proteins that play indispensable roles to suppress the production and signaling of type I and III interferons (IFNs). In this review, we discussed the functions and the underlying mechanisms of different proteins of SARS-CoV-2 that evade the host immune system by suppressing the IFN-β production and TANK-binding kinase 1 (TBK1)/interferon regulatory factor 3 (IRF3)/signal transducer and activator of transcription (STAT)1 and STAT2 phosphorylation. We also described different viral proteins inhibiting the nuclear translocation of IRF3, nuclear factor-κB (NF-κB), and STATs. To date, the following proteins of SARS-CoV-2 including NSP1, NSP6, NSP8, NSP12, NSP13, NSP14, NSP15, open reading frame (ORF)3a, ORF6, ORF8, ORF9b, ORF10, and Membrane (M) protein have been well studied. However, the detailed mechanisms of immune evasion by NSP5, ORF3b, ORF9c, and Nucleocapsid (N) proteins are not well elucidated. Additionally, we also elaborated the perspectives of SARS-CoV-2 proteins.
Keywords: SARS-CoV-2; accessory proteins; immune evasion; non-structural proteins; structural proteins.
Copyright © 2022 Rashid, Xie, Suleman, Shah, Khan and Luo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Evasion of Type I Interferon by SARS-CoV-2.Cell Rep. 2020 Oct 6;33(1):108234. doi: 10.1016/j.celrep.2020.108234. Epub 2020 Sep 19. Cell Rep. 2020. PMID: 32979938 Free PMC article.
-
The role of SARS-CoV-2 accessory proteins in immune evasion.Biomed Pharmacother. 2022 Dec;156:113889. doi: 10.1016/j.biopha.2022.113889. Epub 2022 Oct 17. Biomed Pharmacother. 2022. PMID: 36265309 Free PMC article. Review.
-
Manipulation of innate immune signaling pathways by SARS-CoV-2 non-structural proteins.Front Microbiol. 2022 Nov 21;13:1027015. doi: 10.3389/fmicb.2022.1027015. eCollection 2022. Front Microbiol. 2022. PMID: 36478862 Free PMC article.
-
SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns.Front Immunol. 2021 Jul 7;12:708264. doi: 10.3389/fimmu.2021.708264. eCollection 2021. Front Immunol. 2021. PMID: 34305949 Free PMC article. Review.
-
SARS-CoV-2 NSP13 interacts with host IRF3, blocking antiviral immune responses.J Med Virol. 2023 Jun;95(6):e28881. doi: 10.1002/jmv.28881. J Med Virol. 2023. PMID: 37314155
Cited by
-
Exploring the Binding Effects of Natural Products and Antihypertensive Drugs on SARS-CoV-2: An In Silico Investigation of Main Protease and Spike Protein.Int J Mol Sci. 2023 Nov 2;24(21):15894. doi: 10.3390/ijms242115894. Int J Mol Sci. 2023. PMID: 37958877 Free PMC article.
-
SARS-CoV-2 ORF8 as a Modulator of Cytokine Induction: Evidence and Search for Molecular Mechanisms.Viruses. 2024 Jan 22;16(1):161. doi: 10.3390/v16010161. Viruses. 2024. PMID: 38275971 Free PMC article. Review.
-
Role of interferons in the antiviral battle: from virus-host crosstalk to prophylactic and therapeutic potential in SARS-CoV-2 infection.Front Immunol. 2024 Jan 15;14:1273604. doi: 10.3389/fimmu.2023.1273604. eCollection 2023. Front Immunol. 2024. PMID: 38288121 Free PMC article. Review.
-
Molecular insights into onset of autoimmunity in SARS-CoV-2 infected patients.Rheumatol Autoimmun. 2022 Dec;2(4):198-202. doi: 10.1002/rai2.12056. Epub 2022 Oct 27. Rheumatol Autoimmun. 2022. PMID: 36714799 Free PMC article.
-
Non-coding RNAs derived from the foot-and-mouth disease virus genome trigger broad antiviral activity against coronaviruses.Front Immunol. 2023 Mar 29;14:1166725. doi: 10.3389/fimmu.2023.1166725. eCollection 2023. Front Immunol. 2023. PMID: 37063925 Free PMC article.
References
-
- Pišlar A, Mitrović A, Sabotič J, Pečar Fonović U, Perišić Nanut M, Jakoš T, et al. . The role of cysteine peptidases in coronavirus cell entry and replication: The therapeutic potential of cathepsin inhibitors. PLoS Pathog (2020) 16(11):e1009013. doi: 10.1371/journal.ppat.1009013 - DOI - PMC - PubMed
-
- Siu YL, Teoh KT, Lo J, Chan CM, Kien F, Escriou N, et al. . The m, e, and n structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol (2008) 82(22):11318–30. doi: 10.1128/JVI.01052-08 - DOI - PMC - PubMed
-
- Malik YA. Properties of coronavirus and SARS-CoV-2. Malays J Pathol (2020) 42(1):3–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

