Genotyping by Sequencing Advancements in Barley
- PMID: 36003814
- PMCID: PMC9394214
- DOI: 10.3389/fpls.2022.931423
Genotyping by Sequencing Advancements in Barley
Abstract
Barley is considered an ideal crop to study cereal genetics due to its close relationship with wheat and diploid ancestral genome. It plays a crucial role in reducing risks to global food security posed by climate change. Genetic variations in the traits of interest in crops are vital for their improvement. DNA markers have been widely used to estimate these variations in populations. With the advancements in next-generation sequencing, breeders could access different types of genetic variations within different lines, with single-nucleotide polymorphisms (SNPs) being the most common type. However, genotyping barley with whole genome sequencing (WGS) is challenged by the higher cost and computational demand caused by the large genome size (5.5GB) and a high proportion of repetitive sequences (80%). Genotyping-by-sequencing (GBS) protocols based on restriction enzymes and target enrichment allow a cost-effective SNP discovery by reducing the genome complexity. In general, GBS has opened up new horizons for plant breeding and genetics. Though considered a reliable alternative to WGS, GBS also presents various computational difficulties, but GBS-specific pipelines are designed to overcome these challenges. Moreover, a robust design for GBS can facilitate the imputation to the WGS level of crops with high linkage disequilibrium. The complete exploitation of GBS advancements will pave the way to a better understanding of crop genetics and offer opportunities for the successful improvement of barley and its close relatives.
Keywords: SNPs; barley; genotyping by sequencing; restriction enzymes; target enrichment.
Copyright © 2022 Rajendran, Qureshi and Pourkheirandish.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

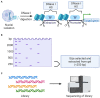
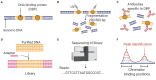
References
-
- Abed A., Légaré G., Pomerleau S., St-Cyr J., Boyle B., Belzile F. J. (2019). Genotyping-by-Sequencing on the Ion Torrent Platform in Barley. New York: Springer New York, 233–252. - PubMed
-
- Babu R., Nair S. K., Prasanna B. M., Gupta H. S. (2004). Integrating marker-assisted selection in crop breeding – prospects and challenges. Curr. Sci. 87, 607–619.
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

