PlaqView 2.0: A comprehensive web portal for cardiovascular single-cell genomics
- PMID: 36003902
- PMCID: PMC9393487
- DOI: 10.3389/fcvm.2022.969421
PlaqView 2.0: A comprehensive web portal for cardiovascular single-cell genomics
Abstract
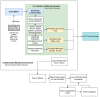
Single-cell RNA-seq (scRNA-seq) is a powerful genomics technology to interrogate the cellular composition and behaviors of complex systems. While the number of scRNA-seq datasets and available computational analysis tools have grown exponentially, there are limited systematic data sharing strategies to allow rapid exploration and re-analysis of single-cell datasets, particularly in the cardiovascular field. We previously introduced PlaqView, an open-source web portal for the exploration and analysis of published atherosclerosis single-cell datasets. Now, we introduce PlaqView 2.0 (www.plaqview.com), which provides expanded features and functionalities as well as additional cardiovascular single-cell datasets. We showcase improved PlaqView functionality, backend data processing, user-interface, and capacity. PlaqView brings new or improved tools to explore scRNA-seq data, including gene query, metadata browser, cell identity prediction, ad hoc RNA-trajectory analysis, and drug-gene interaction prediction. PlaqView serves as one of the largest central repositories for cardiovascular single-cell datasets, which now includes data from human aortic aneurysm, gene-specific mouse knockouts, and healthy references. PlaqView 2.0 brings advanced tools and high-performance computing directly to users without the need for any programming knowledge. Lastly, we outline steps to generalize and repurpose PlaqView's framework for single-cell datasets from other fields.
Keywords: cardiovascular; database; genomics; scRNA-seq; single-cell; web portal.
Copyright © 2022 Ma, Turner, Gancayco, Wong, Song, Mosquera, Auguste, Hodonsky, Prabhakar, Ekiz, van der Laan and Miller.
Conflict of interest statement
Author SL has received Roche funding for unrelated work. The remining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

