Fecal microbial signatures of healthy Han individuals from three bio-geographical zones in Guangdong
- PMID: 36003930
- PMCID: PMC9393523
- DOI: 10.3389/fmicb.2022.920780
Fecal microbial signatures of healthy Han individuals from three bio-geographical zones in Guangdong
Abstract
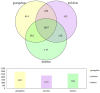
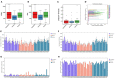
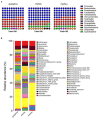
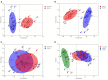
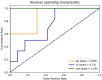
Important forensic evidence traced from crime scenes, such as fecal materials, can help in the forensic investigation of criminal cases. Intestines are the largest microbial pool in the human body whose microbial community is considered to be the human "second fingerprint". The present study explored the potential for community characteristics of gut microbes in forensic medicine. Fecal microbiota profiles of healthy individuals from three representative Han populations (Guangzhou, Shantou and Meizhou) in Guangdong Province, China were evaluated using High-throughput sequencing of V3-V4 hypervariable regions of the 16SrRNA gene. Results of the present study showed that at the genus level, Shantou, Guangzhou, and Meizhou behaved as Enterotype1, Enterotype2, and Enterotype3, which were mainly composed of Bacteroides, Prevotella, and Blautia, respectively. Based on OTU abundance at the genus level, using the random forest prediction model, it was found that there might be potential for distinguishing individuals of Guangzhou, Meizhou, and Shantou according to their fecal microbial community. Moreover, the findings of the microbial community of fecal samples in the present study were significantly different from that of saliva samples reported in our previous study, and thus it is evident that the saliva and feces can be distinguished. In conclusion, this study reported the fecal microbial signature of three Han populations, which may provide basic data for the potential application in forensic practice, containing body fluid identification, and geographical inference.
Keywords: 16S rRNA gene sequencing; Guangdong Han individuals; feces; forensic medicine; gut microbiome.
Copyright © 2022 Huang, Deng, Liu, Huang, Han, Xiao, Liang, Sun, Liu and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Integrating the salivary microbiome in the forensic toolkit by 16S rRNA gene: potential application in body fluid identification and biogeographic inference.Int J Legal Med. 2022 Jul;136(4):975-985. doi: 10.1007/s00414-022-02831-z. Epub 2022 May 10. Int J Legal Med. 2022. PMID: 35536322
-
Skin locations inference and body fluid identification from skin microbial patterns for forensic applications.Forensic Sci Int. 2024 Sep;362:112152. doi: 10.1016/j.forsciint.2024.112152. Epub 2024 Jul 22. Forensic Sci Int. 2024. PMID: 39067177
-
Signatures of vaginal microbiota by 16S rRNA gene: potential bio-geographical application in Chinese Han from three regions of China.Int J Legal Med. 2021 Jul;135(4):1213-1224. doi: 10.1007/s00414-021-02525-y. Epub 2021 Feb 17. Int J Legal Med. 2021. PMID: 33594458
-
Microbial DNA in human nucleic acid extracts: Recoverability of the microbiome in DNA extracts stored frozen long-term and its potential and ethical implications for forensic investigation.Forensic Sci Int Genet. 2022 Jul;59:102686. doi: 10.1016/j.fsigen.2022.102686. Epub 2022 Mar 23. Forensic Sci Int Genet. 2022. PMID: 35338895 Review.
-
Trick or Treating in Forensics-The Challenge of the Saliva Microbiome: A Narrative Review.Microorganisms. 2020 Sep 29;8(10):1501. doi: 10.3390/microorganisms8101501. Microorganisms. 2020. PMID: 33003446 Free PMC article. Review.
Cited by
-
Metagenomics reveals unique gut mycobiome biomarkers in psoriasis.Skin Res Technol. 2024 Jul;30(7):e13822. doi: 10.1111/srt.13822. Skin Res Technol. 2024. PMID: 38970783 Free PMC article.
-
Machine learning integrates region-specific microbial signatures to distinguish geographically adjacent populations within a province.Front Microbiol. 2025 Jul 11;16:1586195. doi: 10.3389/fmicb.2025.1586195. eCollection 2025. Front Microbiol. 2025. PMID: 40718818 Free PMC article.
References
-
- Breiman (2001). Random forests. Mach. Learn. 45, 5–32. 10.1023/A:1010933404324 - DOI
LinkOut - more resources
Full Text Sources

