A comprehensive comparison of fecal microbiota in three ecological bird groups of raptors, waders, and waterfowl
- PMID: 36003944
- PMCID: PMC9393522
- DOI: 10.3389/fmicb.2022.919111
A comprehensive comparison of fecal microbiota in three ecological bird groups of raptors, waders, and waterfowl
Abstract
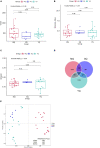
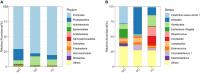
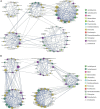
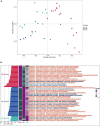
Gut microbiota plays a vital role in maintaining the health and immunity of wild birds. However, less is known about the comparison of fecal microbiota between different ecological groups of wild birds, particularly in the Yellow River National Wetland in Baotou, China, an important transit point for birds migrating all over the East Asia-Australian and Central Asian flyways. In this study, we characterized the fecal microbiota and potential microbial function in nine bird species of raptors, waders, and waterfowl using 16S rRNA gene amplicon sequencing to reveal the microbiota differences and interaction patterns. The results indicated that there was no significant difference in α-diversity, but a significant difference in β-diversity between the three groups of birds. The fecal bacterial microbiota was dominated by Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes in all groups of birds. Furthermore, we identified five bacterial genera that were significantly higher in raptors, five genera that were significantly higher in waders, and two genera that were more abundant in waterfowl. The bacterial co-occurrence network results revealed 15 and 26 key genera in raptors and waterfowls, respectively. The microbial network in waterfowl exhibited a stronger correlation pattern than that in raptors. PICRUSt2 predictions indicated that fecal bacterial function was significantly enriched in the antibiotic biosynthesis pathway in all three groups. Metabolic pathways related to cell motility (bacterial chemotaxis and flagellar assembly) were significantly more abundant in raptors than in waders, whereas waders were enriched in lipid metabolism (synthesis and degradation of ketone bodies and fatty acid biosynthesis). The fecal microbiota in waterfowl harbored more abundant vitamin B6 metabolism, RNA polymerase, and tyrosine and tryptophan biosynthesis. This comparative study revealed the microbial community structure, microbial co-occurrence patterns, and potential functions, providing a better understanding of the ecology and conservation of wild birds. Future studies may focus on unraveling metagenomic functions and dynamics along with the migration routine or different seasons by metagenomics or metatranscriptomics.
Keywords: co-occurrence network; fecal microbiota; metabolic pathways; raptors; waders; waterfowl.
Copyright © 2022 Zhao, Liu, Gao and Bai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Benson A. K., Kelly S. A., Legge R., Ma F., Low S. J., Kim J., et al. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U.S.A. 107 18933–18938. 10.1073/pnas.1007028107 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

