A mosaic-type trimeric RBD-based COVID-19 vaccine candidate induces potent neutralization against Omicron and other SARS-CoV-2 variants
- PMID: 36004719
- PMCID: PMC9481243
- DOI: 10.7554/eLife.78633
A mosaic-type trimeric RBD-based COVID-19 vaccine candidate induces potent neutralization against Omicron and other SARS-CoV-2 variants
Abstract
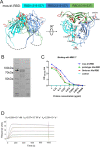
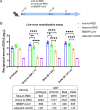
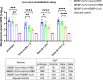
Large-scale populations in the world have been vaccinated with COVID-19 vaccines, however, breakthrough infections of SARS-CoV-2 are still growing rapidly due to the emergence of immune-evasive variants, especially Omicron. It is urgent to develop effective broad-spectrum vaccines to better control the pandemic of these variants. Here, we present a mosaic-type trimeric form of spike receptor-binding domain (mos-tri-RBD) as a broad-spectrum vaccine candidate, which carries the key mutations from Omicron and other circulating variants. Tests in rats showed that the designed mos-tri-RBD, whether used alone or as a booster shot, elicited potent cross-neutralizing antibodies against not only Omicron but also other immune-evasive variants. Neutralizing antibody ID50 titers induced by mos-tri-RBD were substantially higher than those elicited by homo-tri-RBD (containing homologous RBDs from prototype strain) or the BIBP inactivated COVID-19 vaccine (BBIBP-CorV). Our study indicates that mos-tri-RBD is highly immunogenic, which may serve as a broad-spectrum vaccine candidate in combating SARS-CoV-2 variants including Omicron.
Keywords: COVID-19; SARS-CoV-2; broad-spectrum; immunology; infectious disease; inflammation; microbiology; omicron variant; vaccine; viruses.
Plain language summary
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic continues to pose a serious threat to public health and has so far resulted in over six million deaths worldwide. Mass vaccination programs have reduced the risk of serious illness and death in many people, but the virus continues to persist and circulate in communities across the globe. Furthermore, the current vaccines may be less effective against the new variants of the virus, such as Omicron and Delta, which are continually emerging and evolving. Therefore, it is urgent to develop effective vaccines that can provide broad protection against existing and future forms of SARS-CoV-2. There are several different types of SARS-CoV-2 vaccine, but they all work in a similar way. They contain molecules that induce immune responses in individuals to help the body recognize and more effectively fight SARS-CoV-2 if they happen to encounter it in the future. These immune responses may be so specific that new variants of a virus may not be recognized by them. Therefore, a commonly used strategy for producing vaccines with broad protection is to make multiple vaccines that each targets different variants and then mix them together before administering to patients. Here, Zhang et al. took a different approach by designing a new vaccine candidate against SARS-CoV2 that contained three different versions of part of a SARS-CoV2 protein – the so-called spike protein – all linked together as one molecule. The different versions of the spike protein fragment were designed to include key features of the fragments found in Omicron and several other SARS-CoV-2 variants. The experiments found that this candidate vaccine elicited a much higher immune response against Omicron and other SARS-CoV-2 variants in rats than an existing SARS-CoV-2 vaccine. It was also effective as a booster shot after a first vaccination with the existing SARS-CoV-2 vaccine. These findings demonstrate that the molecule developed by Zhang et al. induces potent and broad immune responses against different variants of SARS-CoV-2 including Omicron in rats. The next steps following on from this work are to evaluate the safety and immunogenicity of this vaccine candidate in clinical trials. In the future, it may be possible to use a similar approach to develop new broad-spectrum vaccines against other viruses.
© 2022, Zhang, Han et al.
Conflict of interest statement
JZ, ZH, YL, XZ, YJ, LD, SS, JH, ZL, ZL, YH, NL, FS, JS, QL is listed as an inventor of the pending patent application for the mos-tri-RBD vaccine (Application number: 202210083654.X), HW, JZ is an employee of Beijing Institute of Biological Products Company Limited, KX, WL, JW, XZ, XL, XL, WH, GW No competing interests declared
Figures



Similar articles
-
Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19).2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 34033342 Free Books & Documents.
-
Pan-beta-coronavirus subunit vaccine prevents SARS-CoV-2 Omicron, SARS-CoV, and MERS-CoV challenge.J Virol. 2024 Sep 17;98(9):e0037624. doi: 10.1128/jvi.00376-24. Epub 2024 Aug 27. J Virol. 2024. PMID: 39189731 Free PMC article.
-
A Glycosylated RBD Protein Induces Enhanced Neutralizing Antibodies against Omicron and Other Variants with Improved Protection against SARS-CoV-2 Infection.J Virol. 2022 Sep 14;96(17):e0011822. doi: 10.1128/jvi.00118-22. Epub 2022 Aug 16. J Virol. 2022. PMID: 35972290 Free PMC article.
-
The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Corcern.Front Med (Lausanne). 2022 May 20;9:849217. doi: 10.3389/fmed.2022.849217. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35669924 Free PMC article. Review.
-
Infection-mediated immune response in SARS-CoV-2 breakthrough infection and implications for next-generation COVID-19 vaccine development.Vaccine. 2024 Feb 27;42(6):1401-1406. doi: 10.1016/j.vaccine.2024.01.088. Epub 2024 Feb 2. Vaccine. 2024. PMID: 38310015 Review.
Cited by
-
The Effect of Select SARS-CoV-2 N-Linked Glycan and Variant of Concern Spike Protein Mutations on C-Type Lectin-Receptor-Mediated Infection.Viruses. 2023 Sep 9;15(9):1901. doi: 10.3390/v15091901. Viruses. 2023. PMID: 37766307 Free PMC article.
-
Advances in SARS-CoV-2 receptor-binding domain-based COVID-19 vaccines.Expert Rev Vaccines. 2023 Jan-Dec;22(1):422-439. doi: 10.1080/14760584.2023.2211153. Expert Rev Vaccines. 2023. PMID: 37161869 Free PMC article. Review.
-
Development and characterization of a multimeric recombinant protein using the spike protein receptor binding domain as an antigen to induce SARS-CoV-2 neutralization.Immun Inflamm Dis. 2024 Jul;12(7):e1353. doi: 10.1002/iid3.1353. Immun Inflamm Dis. 2024. PMID: 39056544 Free PMC article.
-
Host-Pathogen Interaction Interface: Promising Candidate Targets for Vaccine-Induced Protective and Memory Immune Responses.Vaccines (Basel). 2025 Apr 16;13(4):418. doi: 10.3390/vaccines13040418. Vaccines (Basel). 2025. PMID: 40333316 Free PMC article. Review.
-
Broad protective RBD heterotrimer vaccines neutralize SARS-CoV-2 including Omicron sub-variants XBB/BQ.1.1/BF.7.PLoS Pathog. 2023 Sep 18;19(9):e1011659. doi: 10.1371/journal.ppat.1011659. eCollection 2023 Sep. PLoS Pathog. 2023. PMID: 37721934 Free PMC article.
References
-
- Barouch DH, O’Brien KL, Simmons NL, King SL, Abbink P, Maxfield LF, Sun Y-H, La Porte A, Riggs AM, Lynch DM, Clark SL, Backus K, Perry JR, Seaman MS, Carville A, Mansfield KG, Szinger JJ, Fischer W, Muldoon M, Korber B. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nature Medicine. 2010;16:319–323. doi: 10.1038/nm.2089. - DOI - PMC - PubMed
-
- Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, Huang W, Li Q, Wang P, An R, Wang J, Wang Y, Niu X, Yang S, Liang H, Sun H, Li T, Yu Y, Cui Q, Liu S, Yang X, Du S, Zhang Z, Hao X, Shao F, Jin R, Wang X, Xiao J, Wang Y, Xie XS. Omicron escapes the majority of existing SARS-cov-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

