1-L Transcription in Alzheimer's Disease
- PMID: 36005139
- PMCID: PMC9406503
- DOI: 10.3390/cimb44080243
1-L Transcription in Alzheimer's Disease
Abstract
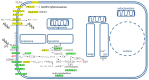
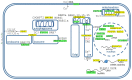
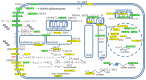
Alzheimer's disease is a very complex disease and better explanations and models are needed to understand how neurons are affected and microglia are activated. A new model of Alzheimer's disease is presented here, the β-amyloid peptide is considered an important RNA recognition/binding peptide. 1-L transcription revealed compatible sequences with AAUAAA (PAS signal) and UUUC (class III ARE rich in U) in the Aβ peptide, supporting the peptide-RNA regulatory model. When a hypothetical model of fibril selection with the prionic character of amyloid assemblies is added to the peptide-RNA regulatory model, the downregulation of the PI3K-Akt pathway and the upregulation of the PLC-IP3 pathway are well explained. The model explains why neurons are less protected from inflammation and why microglia are activated; why mitochondria are destabilized; why the autophagic flux is destabilized; and why the post-transcriptional attenuation of the axonal signal "noise" is interrupted. For example, the model suggests that Aβ peptide may post-transcriptionally control ELAVL2 (ELAV-like RNA binding protein 2) and DCP2 (decapping mRNA protein 2), which are known to regulate RNA processing, transport, and stability.
Keywords: Alzheimer’s disease; bioinformatics method; identified genes; protein–RNA recognition code; β-amyloid peptide.
Conflict of interest statement
The author declares no conflict of interest.
Figures



Similar articles
-
1-L Transcription in Parkinson's Disease.Front Biosci (Landmark Ed). 2023 Nov 23;28(11):292. doi: 10.31083/j.fbl2811292. Front Biosci (Landmark Ed). 2023. PMID: 38062843
-
Precursor-Independent Overproduction of Beta-Amyloid in AD: Mitochondrial Dysfunction as Possible Initiator of Asymmetric RNA-Dependent βAPP mRNA Amplification. An Engine that Drives Alzheimer's Disease.Ann Integr Mol Med. 2019;1(1):61-74. Epub 2019 Nov 20. Ann Integr Mol Med. 2019. PMID: 31858090 Free PMC article.
-
Alzheimer's Disease is Driven by Intraneuronally Retained Beta-Amyloid Produced in the AD-Specific, βAPP-Independent Pathway: Current Perspective and Experimental Models for Tomorrow.Ann Integr Mol Med. 2020;2(1):90-114. doi: 10.33597/aimm.02-1007. Ann Integr Mol Med. 2020. PMID: 32617536 Free PMC article.
-
Neuronal autophagy: self-eating or self-cannibalism in Alzheimer's disease.Neurochem Res. 2013 Sep;38(9):1769-73. doi: 10.1007/s11064-013-1082-4. Epub 2013 Jun 5. Neurochem Res. 2013. PMID: 23737325 Free PMC article. Review.
-
Alzheimer's disease.Subcell Biochem. 2012;65:329-52. doi: 10.1007/978-94-007-5416-4_14. Subcell Biochem. 2012. PMID: 23225010 Review.
Cited by
-
1-L Transcription of SARS-CoV-2 Spike Protein S1 Subunit.Int J Mol Sci. 2024 Apr 18;25(8):4440. doi: 10.3390/ijms25084440. Int J Mol Sci. 2024. PMID: 38674024 Free PMC article.
-
Refining antipsychotic treatment strategies in schizophrenia: discovery of genetic biomarkers for enhanced drug response prediction.Mol Psychiatry. 2025 Jun;30(6):2362-2371. doi: 10.1038/s41380-024-02841-w. Epub 2024 Nov 19. Mol Psychiatry. 2025. PMID: 39562719
-
1-L Transcription in Prion Diseases.Int J Mol Sci. 2024 Sep 15;25(18):9961. doi: 10.3390/ijms25189961. Int J Mol Sci. 2024. PMID: 39337449 Free PMC article.
-
Editorial for the Special Issue "Aging, Age-Related Changes in the Brain and the Progression of Alzheimer's Disease".Curr Issues Mol Biol. 2025 Apr 29;47(5):318. doi: 10.3390/cimb47050318. Curr Issues Mol Biol. 2025. PMID: 40699717 Free PMC article.
-
mosGraphFlow: a novel integrative graph AI model mining disease targets from multi-omic data.bioRxiv [Preprint]. 2024 Sep 3:2024.08.01.606219. doi: 10.1101/2024.08.01.606219. bioRxiv. 2024. PMID: 39282361 Free PMC article. Preprint.
References
-
- Frisoni G.B., Altomare D., Thal D.R., Ribaldi F., van der Kant R., Ossenkoppele R., Blennow K., Cummings J., van Duijn C., Nilsson P.M., et al. The probabilistic model of alzheimer disease: The amyloid hypothesis revised. Nat. Rev. Neurosci. 2022;23:53–66. doi: 10.1038/s41583-021-00533-w. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

