Integrative Multi-Omics Analysis for the Determination of Non-Muscle Invasive vs. Muscle Invasive Bladder Cancer: A Pilot Study
- PMID: 36005168
- PMCID: PMC9406560
- DOI: 10.3390/curroncol29080430
Integrative Multi-Omics Analysis for the Determination of Non-Muscle Invasive vs. Muscle Invasive Bladder Cancer: A Pilot Study
Abstract
Objectives: The molecular landscape of non-muscle-invasive (NMIBC) and muscle-invasive (MIBC) bladder cancer based on molecular characteristics is essential but poorly understood. In this pilot study we aimed to identify a multi-omics signature that can distinguish MIBC from NMIBC. Such a signature can assist in finding potential mechanistic biomarkers and druggable targets.
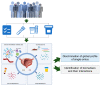
Methods: Patients diagnosed with NMIBC (n = 15) and MIBC (n = 11) were recruited at a tertiary-care hospital in Nanjing from 1 April 2021, and 31 July 2021. Blood, urine and stool samples per participant were collected, in which the serum metabolome, urine metabolome, gut microbiome, and serum extracellular vesicles (EV) proteome were quantified. The differences of the global profiles and individual omics measure between NMIBC vs. MIBC were assessed by permutational multivariate analysis and the Mann-Whitney test, respectively. Logistic regression analysis was used to assess the association of each identified analyte with NMIBC vs. MIBC, and the Spearman correlation was used to investigate the correlations between identified analytes, where both were adjusted for age, sex and smoking status.
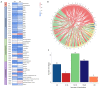
Results: Among 3168 multi-omics measures that passed the quality control, 159 were identified to be differentiated in NMIBC vs. MIBC. Of these, 46 analytes were associated with bladder cancer progression. In addition, the global profiles showed significantly different urine metabolome (p = 0.029), gut microbiome (p = 0.036), and serum EV (extracellular vesicles) proteome (p = 0.039) but not serum metabolome (p = 0.059). We also observed 17 (35%) analytes that had been developed as drug targets. Multiple interactions were obtained between the identified analytes, whereas for the majority (61%), the number of interactions was at 11-20. Moreover, unconjugated bilirubin (p = 0.009) and white blood cell count (p = 0.006) were also shown to be different in NMIBC and MIBC, and associated with 11 identified omics analytes.
Conclusions: The pilot study has shown promising to monitor the progression of bladder cancer by integrating multi-omics data and deserves further investigations.
Keywords: bladder cancer; molecular epidemiology; multi-omics; progression of disease.
Conflict of interest statement
All the authors declare that they have no conflict of interest.
Figures


Similar articles
-
Non-invasive diagnosis and surveillance of bladder cancer with driver and passenger DNA methylation in a prospective cohort study.Clin Transl Med. 2022 Aug;12(8):e1008. doi: 10.1002/ctm2.1008. Clin Transl Med. 2022. PMID: 35968916 Free PMC article.
-
Comparative bioinformatics analysis of prognostic and differentially expressed genes in non-muscle and muscle invasive bladder cancer.J Proteomics. 2020 Oct 30;229:103951. doi: 10.1016/j.jprot.2020.103951. Epub 2020 Aug 27. J Proteomics. 2020. PMID: 32860965
-
Investigating the association between the urinary microbiome and bladder cancer: An exploratory study.Urol Oncol. 2021 Jun;39(6):370.e9-370.e19. doi: 10.1016/j.urolonc.2020.12.011. Epub 2021 Jan 9. Urol Oncol. 2021. PMID: 33436328
-
Prognostic Implications of Immunohistochemical Biomarkers in Non-muscle-invasive Blad Cancer and Muscle-invasive Bladder Cancer.Mini Rev Med Chem. 2020;20(12):1133-1152. doi: 10.2174/1389557516666160512151202. Mini Rev Med Chem. 2020. PMID: 27173513 Review.
-
LC-MS metabolomics of urine reveals distinct profiles for non-muscle-invasive and muscle-invasive bladder cancer.World J Urol. 2022 Oct;40(10):2387-2398. doi: 10.1007/s00345-022-04136-7. Epub 2022 Sep 4. World J Urol. 2022. PMID: 36057894 Review.
Cited by
-
The role of the urinary microbiome in genitourinary cancers.Nat Rev Urol. 2025 Aug;22(8):544-561. doi: 10.1038/s41585-025-01011-z. Epub 2025 Mar 13. Nat Rev Urol. 2025. PMID: 40082677 Review.
-
Unmasking the Metabolite Signature of Bladder Cancer: A Systematic Review.Int J Mol Sci. 2024 Mar 15;25(6):3347. doi: 10.3390/ijms25063347. Int J Mol Sci. 2024. PMID: 38542319 Free PMC article.
-
Serum metabolomic analysis identified serum biomarkers predicting tumour recurrence after Bacillus Calmette-Guérin therapy in patients with non-muscle invasive bladder cancer.Bladder Cancer. 2025 Mar 18;11(1):23523735251325100. doi: 10.1177/23523735251325100. eCollection 2025 Jan-Mar. Bladder Cancer. 2025. PMID: 40109498 Free PMC article.
-
Emerging RNA-Based Therapeutic and Diagnostic Options: Recent Advances and Future Challenges in Genitourinary Cancers.Int J Mol Sci. 2023 Feb 27;24(5):4601. doi: 10.3390/ijms24054601. Int J Mol Sci. 2023. PMID: 36902032 Free PMC article. Review.
-
Applications of Exosomes in Diagnosing Muscle Invasive Bladder Cancer.Pharmaceutics. 2022 Sep 23;14(10):2027. doi: 10.3390/pharmaceutics14102027. Pharmaceutics. 2022. PMID: 36297462 Free PMC article. Review.
References
-
- Abdollah F., Gandaglia G., Thuret R., Schmitges J., Tian Z., Jeldres C., Passoni N.M., Briganti A., Shariat S.F., Perrotte P., et al. Incidence, survival and mortality rates of stage-specific bladder cancer in United States: A trend analysis. Cancer Epidemiology. 2013;37:219–225. doi: 10.1016/j.canep.2013.02.002. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

