Differences in Supragingival Microbiome in Patients with and without Full-Crown Prostheses
- PMID: 36005250
- PMCID: PMC9406617
- DOI: 10.3390/dj10080152
Differences in Supragingival Microbiome in Patients with and without Full-Crown Prostheses
Abstract
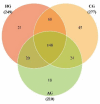
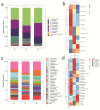
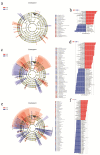
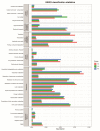
Objectives: To characterize the microflora profile of supragingival biofilm in patients with and without full-crown prostheses. Methods: Plaque samples of full-crown prostheses and teeth in patients with porcelain-fused-to-metal crowns, all-ceramic crowns, and no prostheses were collected (three patients per group), using 16S rRNA high-throughput sequencing technology to conduct DNA sequencing on the samples and using Qiime, R, and PICRUSt2 software to perform bioinformatics analyses and functional analyses on sequencing data. Results: In total, 110,209 valid sequences were obtained in the experiment, corresponding to 11 phyla and 120 genera. The predominant species shared by the three groups were phyla Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria and genera Rothia, Porphyromonas, Prevotella, Streptococcus, Veillonella, Leptotrichia, Neisseria, Citrobacter, and Pseudomonas. The species-difference analysis showed that genus Hameophilus significantly increased after the patient wore the dental prosthesis. Compared with the no-prosthesis samples, the functional analysis showed that cell motility increased in the samples from full-crown prostheses, while replication and repair, and translation decreased. Conclusions: This study reveals the changes in the oral microbial community of patients with full-crown prostheses, which could provide insights regarding the safety of materials for long-term use in the oral cavity.
Keywords: 16S rRNA high-throughput sequencing; community structure; full-crown prosthesis; oral microorganism.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Microbial community analysis of supragingival plaque in patients with fixed prostheses.J Prosthet Dent. 2025 Jan 6:S0022-3913(24)00832-1. doi: 10.1016/j.prosdent.2024.12.015. Online ahead of print. J Prosthet Dent. 2025. PMID: 39765413
-
Plaque bacterial microbiome diversity in children younger than 30 months with or without caries prior to eruption of second primary molars.PLoS One. 2014 Feb 28;9(2):e89269. doi: 10.1371/journal.pone.0089269. eCollection 2014. PLoS One. 2014. PMID: 24586647 Free PMC article.
-
Microbial Similarity and Preference for Specific Sites in Healthy Oral Cavity and Esophagus.Front Microbiol. 2018 Jul 17;9:1603. doi: 10.3389/fmicb.2018.01603. eCollection 2018. Front Microbiol. 2018. PMID: 30065718 Free PMC article.
-
Bacterial Diversity and Community Structure of Supragingival Plaques in Adults with Dental Health or Caries Revealed by 16S Pyrosequencing.Front Microbiol. 2016 Jul 22;7:1145. doi: 10.3389/fmicb.2016.01145. eCollection 2016. Front Microbiol. 2016. PMID: 27499752 Free PMC article.
-
Deciphering Endodontic Microbial Communities by Next-generation Sequencing.J Endod. 2018 Jul;44(7):1080-1087. doi: 10.1016/j.joen.2018.04.003. Epub 2018 Jun 1. J Endod. 2018. PMID: 29861065 Review.
Cited by
-
Isolation of Clinical Microbial Isolates during Orthodontic Aligner Therapy and Their Ability to Form Biofilm.Dent J (Basel). 2023 Jan 3;11(1):13. doi: 10.3390/dj11010013. Dent J (Basel). 2023. PMID: 36661550 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

