Extension of the CHARMM Classical Drude Polarizable Force Field to N- and O-Linked Glycopeptides and Glycoproteins
- PMID: 36005290
- PMCID: PMC9463114
- DOI: 10.1021/acs.jpcb.2c04245
Extension of the CHARMM Classical Drude Polarizable Force Field to N- and O-Linked Glycopeptides and Glycoproteins
Abstract
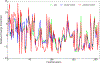
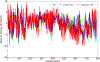
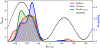
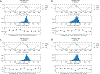
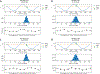
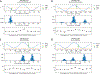
Molecular dynamic simulations are an effective tool to study complex molecular systems and are contingent upon the availability of an accurate and reliable molecular mechanics force field. The Drude polarizable force field, which allows for the explicit treatment of electronic polarization in a computationally efficient fashion, has been shown to reproduce experimental properties that were difficult or impossible to reproduce with the CHARMM additive force field, including peptide folding cooperativity, RNA hairpin structures, and DNA base flipping. Glycoproteins are essential components of glycoconjugate vaccines, antibodies, and many pharmaceutically important molecules, and an accurate polarizable force field that includes compatibility between the protein and carbohydrate aspect of the force field is essential to study these types of systems. In this work, we present an extension of the Drude polarizable force field to glycoproteins, including both N- and O-linked species. Parameter optimization focused on the dihedral terms using a reweighting protocol targeting NMR solution J-coupling data for model glycopeptides. Validation of the model include eight model glycopeptides and four glycoproteins with multiple N- and O-linked glycosylations. The new glycoprotein carbohydrate force field can be used in conjunction with the remainder of Drude polarizable force field through a variety of MD simulation programs including GROMACS, OPENMM, NAMD, and CHARMM and may be accessed through the Drude Prepper module in the CHARMM-GUI.
Conflict of interest statement
The authors declare the following competing interest(s): ADM Jr. is cofounder and CSO of SilcsBio LLC.
Figures

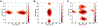
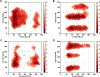
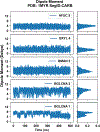
References
-
- Whitford PC; Blanchard SC; Cate JH; Sanbonmatsu KY Connecting the kinetics and energy landscape of tRNA translocation on the ribosome. PLoS Comput Biol 2013, 9 (3), e1003003. DOI: 10.1371/journal.pcbi.1003003. - DOI - PMC - PubMed
- Dror RO; Pan AC; Arlow DH; Borhani DW; Maragakis P; Shan Y; Xu H; Shaw DE Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc Natl Acad Sci U S A 2011, 108 (32), 13118–13123. DOI: 10.1073/pnas.1104614108. - DOI - PMC - PubMed
- Shaw DE; Maragakis P; Lindorff-Larsen K; Piana S; Dror RO; Eastwood MP; Bank JA; Jumper JM; Salmon JK; Shan Y; et al. Atomic-level characterization of the structural dynamics of proteins. Science 2010, 330 (6002), 341–346. DOI: 10.1126/science.1187409. - DOI - PubMed
- McCammon JA; Gelin BR; Karplus M Dynamics of folded proteins. Nature 1977, 267 (5612), 585–590. DOI: 10.1038/267585a0. - DOI - PubMed
-
- Fadda E Molecular Simulations of complex carbohydrates and glycoconjugates. Curr. Opin. Chem. Biology 2022, 69, 102175. - PubMed
-
- Cornell WD; Cieplak P; Bayly CI; Gould IR; Merz KM; Ferguson DM; Spellmeyer DC; Fox T; Caldwell JW; Kollman PA A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. Journal of the American Chemical Society 1995, 117 (19), 5179–5197. DOI: 10.1021/ja00124a002. - DOI
- MacKerell AD; Bashford D; Bellott M; Dunbrack RL; Evanseck JD; Field MJ; Fischer S; Gao J; Guo H; Ha S; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 1998, 102 (18), 3586–3616. DOI: 10.1021/jp973084f. - DOI - PubMed
- MacKerell AD; Brooks B; Brooks CL; Nilsson L; Roux B; Won Y; Karplus M CHARMM: The Energy Function and Its Parameterization. In Encyclopedia of Computational Chemistry, John Wiley & Sons, Ltd, 2002.
- MacKerell AD Jr.; Banavali N; Foloppe N Development and current status of the CHARMM force field for nucleic acids. Biopolymers 2000, 56 (4), 257–265. DOI: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. - DOI - PubMed
- Pastor RW; Mackerell AD Jr. Development of the CHARMM Force Field for Lipids. J Phys Chem Lett 2011, 2 (13), 1526–1532. DOI: 10.1021/jz200167q. - DOI - PMC - PubMed
- Kaminski GA; Friesner RA; Tirado-Rives J; Jorgensen WL Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. The Journal of Physical Chemistry B 2001, 105 (28), 6474–6487. DOI: 10.1021/jp003919d. - DOI
- Damm W; Frontera A; Tirado–Rives J; Jorgensen WL OPLS all-atom force field for carbohydrates. Journal of Computational Chemistry 1997, 18 (16), 1955–1970. DOI: 10.1002/(SICI)1096-987X(199712)18:16<1955::AID-JCC1>3.0.CO;2-L (acccessed 2020/01/13). - DOI
- Soares TA; Hunenberger PH; Kastenholz MA; Krautler V; Lenz T; Lins RD; Oostenbrink C; van Gunsteren WF An improved nucleic acid parameter set for the GROMOS force field. J Comput Chem 2005, 26 (7), 725–737. DOI: 10.1002/jcc.20193. - DOI - PubMed
- Pol-Fachin L; Rusu VH; Verli H; Lins RD GROMOS 53A6GLYC, an Improved GROMOS Force Field for Hexopyranose-Based Carbohydrates. J Chem Theory Comput 2012, 8 (11), 4681–4690. DOI: 10.1021/ct300479h. - DOI - PubMed
-
- Shi Y; Xia Z; Zhang J; Best R; Wu C; Ponder JW; Ren P The Polarizable Atomic Multipole-based AMOEBA Force Field for Proteins. J Chem Theory Comput 2013, 9 (9), 4046–4063. DOI: 10.1021/ct4003702. - DOI - PMC - PubMed
- Patel S; Brooks CL III CHARMM Fluctuating Charge Force Fields for Proteins: I Paramaterization and application to bulk organic liquid simulations. J Comp Chem 2004, 25 (1), 1–16. - PubMed
- Wang ZX; Zhang W; Wu C; Lei H; Cieplak P; Duan Y Strike a balance: optimization of backbone torsion parameters of AMBER polarizable force field for simulations of proteins and peptides. J Comput Chem 2006, 27 (6), 781–790. DOI: 10.1002/jcc.20386. - DOI - PMC - PubMed
- Halgren TA; Damm W Polarizable force fields. Current Opinion in Structural Biology 2001, 11 (2), 236–242. DOI: 10.1016/S0959-440X(00)00196-2. - DOI - PubMed
- Lemkul JA; Huang J; Roux B; MacKerell AD Jr. An Empirical Polarizable Force Field Based on the Classical Drude Oscillator Model: Development History and Recent Applications. Chem Rev 2016, 116 (9), 4983–5013. DOI: 10.1021/acs.chemrev.5b00505. - DOI - PMC - PubMed
- Cieplak P; Dupradeau FY; Duan Y; Wang J Polarization effects in molecular mechanical force fields. J Phys Condens Matter 2009, 21 (33), 333102. DOI: 10.1088/0953-8984/21/33/333102. - DOI - PMC - PubMed
- Ponder JW; Case DA Force fields for protein simulations. Advances in Protein Chemistry 2003, 66, 27–85. - PubMed
- Baker CM Polarizable force fields for molecular dynamics simulations of biomolecules. Wiley Interdisciplinary Reviews: Computational Molecular Science 2015, 5 (2), 241–254. DOI: 10.1002/wcms.1215 (acccessed 2020/01/15). - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

