In situ single particle classification reveals distinct 60S maturation intermediates in cells
- PMID: 36005291
- PMCID: PMC9444246
- DOI: 10.7554/eLife.79272
In situ single particle classification reveals distinct 60S maturation intermediates in cells
Abstract
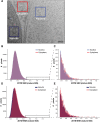
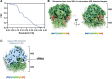
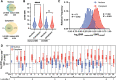
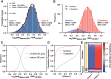
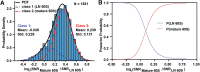
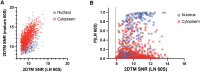
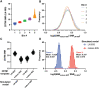
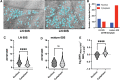
Previously, we showed that high-resolution template matching can localize ribosomes in two-dimensional electron cryo-microscopy (cryo-EM) images of untilted Mycoplasma pneumoniae cells with high precision (Lucas et al., 2021). Here, we show that comparing the signal-to-noise ratio (SNR) observed with 2DTM using different templates relative to the same cellular target can correct for local variation in noise and differentiate related complexes in focused ion beam (FIB)-milled cell sections. We use a maximum likelihood approach to define the probability of each particle belonging to each class, thereby establishing a statistic to describe the confidence of our classification. We apply this method in two contexts to locate and classify related intermediate states of 60S ribosome biogenesis in the Saccharomyces cerevisiae cell nucleus. In the first, we separate the nuclear pre-60S population from the cytoplasmic mature 60S population, using the subcellular localization to validate assignment. In the second, we show that relative 2DTM SNRs can be used to separate mixed populations of nuclear pre-60S that are not visually separable. 2DTM can distinguish related molecular populations without the need to generate 3D reconstructions from the data to be classified, permitting classification even when only a few target particles exist in a cell.
Keywords: 2DTM; FIB-milling; S. cerevisiae; cell biology; in situ cryo-EM; molecular biophysics; nucleus; particle classification; ribosome biogenesis; structural biology.
© 2022, Lucas et al.
Conflict of interest statement
BL, KZ, SL No competing interests declared, NG Reviewing editor, eLife
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

