matRadiomics: A Novel and Complete Radiomics Framework, from Image Visualization to Predictive Model
- PMID: 36005464
- PMCID: PMC9410206
- DOI: 10.3390/jimaging8080221
matRadiomics: A Novel and Complete Radiomics Framework, from Image Visualization to Predictive Model
Abstract
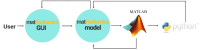
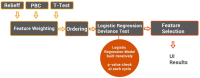
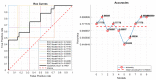
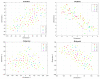
Radiomics aims to support clinical decisions through its workflow, which is divided into: (i) target identification and segmentation, (ii) feature extraction, (iii) feature selection, and (iv) model fitting. Many radiomics tools were developed to fulfill the steps mentioned above. However, to date, users must switch different software to complete the radiomics workflow. To address this issue, we developed a new free and user-friendly radiomics framework, namely matRadiomics, which allows the user: (i) to import and inspect biomedical images, (ii) to identify and segment the target, (iii) to extract the features, (iv) to reduce and select them, and (v) to build a predictive model using machine learning algorithms. As a result, biomedical images can be visualized and segmented and, through the integration of Pyradiomics into matRadiomics, radiomic features can be extracted. These features can be selected using a hybrid descriptive-inferential method, and, consequently, used to train three different classifiers: linear discriminant analysis, k-nearest neighbors, and support vector machines. Model validation is performed using k-fold cross-Validation and k-fold stratified cross-validation. Finally, the performance metrics of each model are shown in the graphical interface of matRadiomics. In this study, we discuss the workflow, architecture, application, future development of matRadiomics, and demonstrate its working principles in a real case study with the aim of establishing a reference standard for the whole radiomics analysis, starting from the image visualization up to the predictive model implementation.
Keywords: CT; MRI; PET; image analysis; machine learning; radiomics; software package.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






References
-
- Comelli A., Stefano A., Coronnello C., Russo G., Vernuccio F., Cannella R., Salvaggio G., Lagalla R., Barone S. Communications in Computer and Information Science, Proceedings of the Medical Image Understanding and Analysis, Oxford, UK, 15–17 July 2020. Volume 1248. Springer; Cham, Switzerland: 2020. Radiomics: A New Biomedical Workflow to Create a Predictive Model; pp. 280–293.
-
- Laudicella R., Comelli A., Liberini V., Vento A., Stefano A., Spataro A., Crocè L., Baldari S., Bambaci M., Deandreis D., et al. [68 Ga]DOTATOC PET/CT Radiomics to Predict the Response in GEP-NETs Undergoing [177 Lu]DOTATOC PRRT: The “Theragnomics” Concept. Cancers. 2022;14:984. doi: 10.3390/cancers14040984. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

