Host-Gut Microbiome Metabolic Interactions in PFAS-Impacted Freshwater Turtles (Emydura macquarii macquarii)
- PMID: 36005619
- PMCID: PMC9415956
- DOI: 10.3390/metabo12080747
Host-Gut Microbiome Metabolic Interactions in PFAS-Impacted Freshwater Turtles (Emydura macquarii macquarii)
Abstract
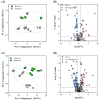
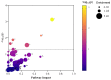
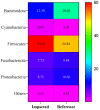
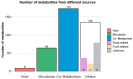
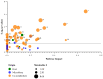
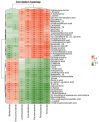
Per-and polyfluoroalkyl substances (PFAS) are a growing concern for humans, wildlife, and more broadly, ecosystem health. Previously, we characterised the microbial and biochemical impact of elevated PFAS on the gut microbiome of freshwater turtles (Emydura macquarii macquarii) within a contaminated catchment in Queensland, Australia. However, the understanding of PFAS impacts on this species and other aquatic organisms is still very limited, especially at the host-gut microbiome molecular interaction level. To this end, the present study aimed to apply these leading-edge omics technologies within an integrated framework that provides biological insight into the host turtle-turtle gut microbiome interactions of PFAS-impacted wild-caught freshwater turtles. For this purpose, faecal samples from PFAS-impacted turtles (n = 5) and suitable PFAS-free reference turtles (n = 5) were collected and analysed. Data from 16S rRNA gene amplicon sequencing and metabolomic profiling of the turtle faeces were integrated using MetOrigin to assign host, microbiome, and co-metabolism activities. Significant variation in microbial composition was observed between the two turtle groups. The PFAS-impacted turtles showed a higher relative abundance of Firmicutes and a lower relative abundance of Bacteroidota than the reference turtles. The faecal metabolome showed several metabolites and pathways significantly affected by PFAS exposure. Turtles exposed to PFAS displayed altered amino acid and butanoate metabolisms, as well as altered purine and pyrimidine metabolism. It is predicted from this study that PFAS-impacted both the metabolism of the host turtle and its gut microbiota which in turn has the potential to influence the host's physiology and health.
Keywords: Emydura macquarii; PFAS; freshwater turtles; metabolomics; microbiome; omics; water pollutants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Georges A. Setting conservation priorities for Australian freshwater turtles. In: Lunney D., Ayers D., editors. Herpetology in Australia: A Diverse Discipline. Royal Zoological Society of New South Wales; Sydney, Australia: 1993. pp. 49–58.
-
- Georges A., Thomson S. Diversity of Australasian freshwater turtles, with an annotated synonymy and keys to species. Zootaxa. 2010;2496:1–37. doi: 10.11646/zootaxa.2496.1.1. - DOI
-
- Government Q. Species Profile—Emydura macquarii macquarii (Murray Turtle) [(accessed on 10 May 2022)];2022 Available online: https://apps.des.qld.gov.au/species-search/details/?id=43.
-
- Chessman B.C. Declines of freshwater turtles associated with climatic drying in Australia’s Murray–Darling Basin. Wildl. Res. 2011;38:664–671. doi: 10.1071/WR11108. - DOI
LinkOut - more resources
Full Text Sources

