Multiple-Site SUMOylation of FMDV 3C Protease and Its Negative Role in Viral Replication
- PMID: 36005757
- PMCID: PMC9472655
- DOI: 10.1128/jvi.00612-22
Multiple-Site SUMOylation of FMDV 3C Protease and Its Negative Role in Viral Replication
Abstract
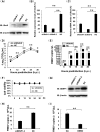
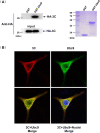
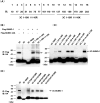
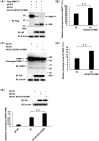
Protein SUMOylation represents an important cellular process that regulates the activities of numerous host proteins as well as of many invasive viral proteins. Foot-and-mouth disease virus (FMDV) is the first animal virus discovered. However, whether SUMOylation takes place during FMDV infection and what role it plays in FMDV pathogenesis have not been investigated. In the present study, we demonstrated that SUMOylation suppressed FMDV replication by small interfering RNA (siRNA) transfection coupled with pharmaceutical inhibition of SUMOylation, which was further confirmed by increased virus replication for SUMOylation-deficient FMDV with mutations in 3C protease, a target of SUMOylation. Moreover, we provided evidence that four lysine residues, Lys-51, -54, -110, and -159, worked together to confer the SUMOylation to the FMDV 3C protease, which may make SUMOylation of FMDV 3C more stable and improve the host's chance of suppressing the replication of FMDV. This is the first report that four lysine residues can be alternatively modified by SUMOylation. Finally, we showed that SUMOylation attenuated the cleavage ability, the inhibitory effect of the interferon signaling pathway, and the protein stability of FMDV 3C, which appeared to correlate with a decrease in FMDV replication. Taken together, the results of our experiments describe a novel cellular regulatory event that significantly restricts FMDV replication through the SUMOylation of 3C protease. IMPORTANCE FMD is a highly contagious and economically important disease in cloven-hoofed animals. SUMOylation, the covalent linkage of a small ubiquitin-like protein to a variety of substrate proteins, has emerged as an important posttranslational modification that plays multiple roles in diverse biological processes. In this study, four lysine residues of FMDV 3C were found to be alternatively modified by SUMOylation. In addition, we demonstrated that SUMOylation attenuated FMDV 3C function through multiple mechanisms, including cleavage ability, the inhibitory effect of the interferon signaling pathway, and protein stability, which, in turn, resulted in a decrease of FMDV replication. Our findings indicate that SUMOylation of FMDV 3C serves as a host cell defense against FMDV replication. Further understanding of the cellular and molecular mechanisms driving this process should offer novel insights to design an effective strategy to control the dissemination of FMDV in animals.
Keywords: 3C protease; SUMOylation; foot-and-mouth disease virus (FMDV); replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

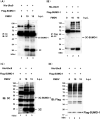

Similar articles
-
Potent small molecule inhibitors against the 3C protease of foot-and-mouth disease virus.Microbiol Spectr. 2024 Apr 2;12(4):e0337223. doi: 10.1128/spectrum.03372-23. Epub 2024 Mar 11. Microbiol Spectr. 2024. PMID: 38466127 Free PMC article.
-
Foot-and-mouth disease virus (FMDV) negatively regulates ZFP36 protein expression to alleviate its antiviral activity.J Virol. 2024 Sep 17;98(9):e0111424. doi: 10.1128/jvi.01114-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194213 Free PMC article.
-
Foot-and-Mouth Disease Virus Antagonizes NOD2-Mediated Antiviral Effects by Inhibiting NOD2 Protein Expression.J Virol. 2019 May 15;93(11):e00124-19. doi: 10.1128/JVI.00124-19. Print 2019 Jun 1. J Virol. 2019. PMID: 30894473 Free PMC article.
-
Foot-and-mouth disease virus 3C protease: recent structural and functional insights into an antiviral target.Int J Biochem Cell Biol. 2007;39(1):1-6. doi: 10.1016/j.biocel.2006.07.006. Epub 2006 Aug 14. Int J Biochem Cell Biol. 2007. PMID: 16979372 Free PMC article. Review.
-
Virus-Host Interactions in Foot-and-Mouth Disease Virus Infection.Front Immunol. 2021 Feb 26;12:571509. doi: 10.3389/fimmu.2021.571509. eCollection 2021. Front Immunol. 2021. PMID: 33717061 Free PMC article. Review.
Cited by
-
The Strategies Used by Animal Viruses to Antagonize Host Antiviral Innate Immunity: New Clues for Developing Live Attenuated Vaccines (LAVs).Vaccines (Basel). 2025 Jan 8;13(1):46. doi: 10.3390/vaccines13010046. Vaccines (Basel). 2025. PMID: 39852825 Free PMC article. Review.
-
SENP1 mediates zinc-induced ZnT6 deSUMOylation at Lys-409 involved in the regulation of zinc metabolism in Golgi apparatus.Cell Mol Life Sci. 2024 Oct 5;81(1):422. doi: 10.1007/s00018-024-05452-4. Cell Mol Life Sci. 2024. PMID: 39367979 Free PMC article.
-
Foot-and-Mouth Disease Virus Capsid Protein VP1 Antagonizes Type I Interferon Signaling via Degradation of Histone Deacetylase 5.Cells. 2024 Mar 19;13(6):539. doi: 10.3390/cells13060539. Cells. 2024. PMID: 38534383 Free PMC article.
-
Protein-S-nitrosylation of human cytomegalovirus pp65 reduces its ability to undermine cGAS.J Virol. 2025 May 20;99(5):e0048125. doi: 10.1128/jvi.00481-25. Epub 2025 Apr 17. J Virol. 2025. PMID: 40243337 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

