Laser Microdissection-Mediated Isolation of Butterfly Wing Tissue for Spatial Transcriptomics
- PMID: 36005768
- PMCID: PMC9415384
- DOI: 10.3390/mps5040067
Laser Microdissection-Mediated Isolation of Butterfly Wing Tissue for Spatial Transcriptomics
Abstract
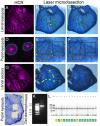
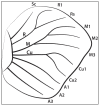
The assignment of specific patterns of gene expression to specific cells in a complex tissue facilitates the connection between genotype and phenotype. Single-cell sequencing of whole tissues produces single-cell transcript resolution but lacks the spatial information of the derivation of each cell, whereas techniques such as multiplex FISH localize transcripts to specific cells in a tissue but require a priori information of the target transcripts to examine. Laser dissection of tissues followed by transcriptome analysis is an efficient and cost-effective technique that provides both unbiased gene expression discovery together with spatial information. Here, we detail a laser dissection protocol for total RNA extraction from butterfly larval and pupal wing tissues, without the need of paraffin embedding or the use of a microtome, that could be useful to researchers interested in the transcriptome of specific areas of the wing during development. This protocol can bypass difficulties in extracting high quality RNA from thick fixed tissues for sequencing applications.
Keywords: Bicyclus anynana; RNAseq; eyespots; laser microdissection; spatial transcriptomics; wing sectors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Dissection of Larval and Pupal Wings of Bicyclus anynana Butterflies.Methods Protoc. 2020 Jan 10;3(1):5. doi: 10.3390/mps3010005. Methods Protoc. 2020. PMID: 31936719 Free PMC article.
-
In situ protocol for butterfly pupal wings using riboprobes.J Vis Exp. 2007;(4):208. doi: 10.3791/208. Epub 2007 May 28. J Vis Exp. 2007. PMID: 18979012 Free PMC article.
-
Cell Dissociation from Butterfly Pupal Wing Tissues for Single-Cell RNA Sequencing.Methods Protoc. 2020 Oct 28;3(4):72. doi: 10.3390/mps3040072. Methods Protoc. 2020. PMID: 33126499 Free PMC article.
-
Advances in isolation and characterization of homogeneous cell populations using laser microdissection.Histol Histopathol. 2005 Jan;20(1):139-46. doi: 10.14670/HH-20.139. Histol Histopathol. 2005. PMID: 15578433 Review.
-
Complementary techniques: laser capture microdissection--increasing specificity of gene expression profiling of cancer specimens.Adv Exp Med Biol. 2007;593:54-65. doi: 10.1007/978-0-387-39978-2_6. Adv Exp Med Biol. 2007. PMID: 17265716 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

