Conserved FimK Truncation Coincides with Increased Expression of Type 3 Fimbriae and Cultured Bladder Epithelial Cell Association in Klebsiella quasipneumoniae
- PMID: 36005809
- PMCID: PMC9487511
- DOI: 10.1128/jb.00172-22
Conserved FimK Truncation Coincides with Increased Expression of Type 3 Fimbriae and Cultured Bladder Epithelial Cell Association in Klebsiella quasipneumoniae
Abstract
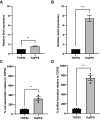
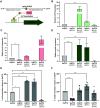
Klebsiella spp. commonly cause both uncomplicated urinary tract infection (UTI) and recurrent UTI (rUTI). Klebsiella quasipneumoniae, a relatively newly defined species of Klebsiella, has been shown to be metabolically distinct from Klebsiella pneumoniae, but its type 1 and type 3 fimbriae have not been studied. K. pneumoniae uses both type 1 and type 3 fimbriae to attach to host epithelial cells. The type 1 fimbrial operon is well conserved between Escherichia coli and K. pneumoniae apart from fimK, which is unique to Klebsiella spp. FimK contains an N-terminal DNA binding domain and a C-terminal phosphodiesterase (PDE) domain that has been hypothesized to cross-regulate type 3 fimbriae expression via modulation of cellular levels of cyclic di-GMP. Here, we find that a conserved premature stop codon in K. quasipneumoniae fimK results in truncation of the C-terminal PDE domain and that K quasipneumoniae strain KqPF9 cultured bladder epithelial cell association and invasion are dependent on type 3 but not type 1 fimbriae. Further, we show that basal expression of both type 1 and type 3 fimbrial operons as well as cultured bladder epithelial cell association is elevated in KqPF9 relative to uropathogenic K. pneumoniae TOP52. Finally, we show that complementation of KqPF9ΔfimK with the TOP52 fimK allele reduced type 3 fimbrial expression and cultured bladder epithelial cell attachment. Taken together these data suggest that the C-terminal PDE of FimK can modulate type 3 fimbrial expression in K. pneumoniae and its absence in K. quasipneumoniae may lead to a loss of type 3 fimbrial cross-regulation. IMPORTANCE K. quasipneumoniae is often indicated as the cause of opportunistic infections, including urinary tract infection, which affects >50% of women worldwide. However, the virulence factors of K. quasipneumoniae remain uninvestigated. Prior to this work, K. quasipneumoniae and K. pneumoniae had only been distinguished phenotypically by metabolic differences. This work contributes to the understanding of K. quasipneumoniae by evaluating the contribution of type 1 and type 3 fimbriae, which are critical colonization factors encoded by all Klebsiella spp., to K. quasipneumoniae bladder epithelial cell attachment in vitro. We observe clear differences in bladder epithelial cell attachment and regulation of type 3 fimbriae between uropathogenic K. pneumoniae and K. quasipneumoniae that coincide with a structural difference in the fimbrial regulatory gene fimK.
Keywords: Klebsiella; Klebsiella pneumoniae; Klebsiella quasipneumoniae; fimbriae; urinary tract infection; urothelium; virulence regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
FimK regulation on the expression of type 1 fimbriae in Klebsiella pneumoniae CG43S3.Microbiology (Reading). 2013 Jul;159(Pt 7):1402-1415. doi: 10.1099/mic.0.067793-0. Epub 2013 May 23. Microbiology (Reading). 2013. PMID: 23704787
-
Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression.Infect Immun. 2008 Jul;76(7):3337-45. doi: 10.1128/IAI.00090-08. Epub 2008 Apr 14. Infect Immun. 2008. PMID: 18411285 Free PMC article.
-
Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence.Infect Immun. 2008 Sep;76(9):4055-65. doi: 10.1128/IAI.00494-08. Epub 2008 Jun 16. Infect Immun. 2008. PMID: 18559432 Free PMC article.
-
Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation.Future Microbiol. 2012 Aug;7(8):991-1002. doi: 10.2217/fmb.12.74. Future Microbiol. 2012. PMID: 22913357 Review.
-
[Research advances in the virulence factors of Klebsiella pneumoniae--A review].Wei Sheng Wu Xue Bao. 2015 Oct 4;55(10):1245-52. Wei Sheng Wu Xue Bao. 2015. PMID: 26939452 Review. Chinese.
Cited by
-
Mapping Antimicrobial Resistance in Escherichia coli and Klebsiella pneumoniae from Complicated Urinary Tract Infections in Oman: Phenotypic and Genotypic Insights.Diagnostics (Basel). 2025 Apr 22;15(9):1062. doi: 10.3390/diagnostics15091062. Diagnostics (Basel). 2025. PMID: 40361883 Free PMC article.
-
Emergence of NDM-7-Producing Klebsiella quasipneumoniae subs. simillipneumoniae ST138 in a Hospital from the Northern Region of Brazil.Microorganisms. 2025 Feb 1;13(2):314. doi: 10.3390/microorganisms13020314. Microorganisms. 2025. PMID: 40005681 Free PMC article.
-
Fluorescent molecular probe for in vivo and in vitro targeting and imaging of an intracellular bacterial infection.Chem Sci. 2025 Mar 24;16(18):7902-7911. doi: 10.1039/d4sc05680a. eCollection 2025 May 7. Chem Sci. 2025. PMID: 40191126 Free PMC article.
References
-
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 112:E3574–81. 10.1073/pnas.1501049112. - DOI - PMC - PubMed
-
- Venkitapathi S, Sharon BM, Ratna TA, Arute AP, Zimmern PE, De Nisco NJ. 2021. Complete genome sequences of three uropathogenic Klebsiella quasipneumoniae strains isolated from postmenopausal women with recurrent urinary tract infection. Microbiol Resour Announc 10:e00073-21. 10.1128/MRA.00073-21. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

