Comparison of Different Label-Free Raman Spectroscopy Approaches for the Discrimination of Clinical MRSA and MSSA Isolates
- PMID: 36005817
- PMCID: PMC9603629
- DOI: 10.1128/spectrum.00763-22
Comparison of Different Label-Free Raman Spectroscopy Approaches for the Discrimination of Clinical MRSA and MSSA Isolates
Abstract
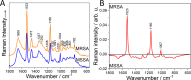
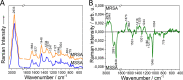
Methicillin-resistant Staphylococcus aureus (MRSA) is classified as one of the priority pathogens that threaten human health. Resistance detection with conventional microbiological methods takes several days, forcing physicians to administer empirical antimicrobial treatment that is not always appropriate. A need exists for a rapid, accurate, and cost-effective method that allows targeted antimicrobial therapy in limited time. In this pilot study, we investigate the efficacy of three different label-free Raman spectroscopic approaches to differentiate methicillin-resistant and -susceptible clinical isolates of S. aureus (MSSA). Single-cell analysis using 532 nm excitation was shown to be the most suitable approach since it captures information on the overall biochemical composition of the bacteria, predicting 87.5% of the strains correctly. UV resonance Raman microspectroscopy provided a balanced accuracy of 62.5% and was not sensitive enough in discriminating MRSA from MSSA. Excitation of 785 nm directly on the petri dish provided a balanced accuracy of 87.5%. However, the difference between the strains was derived from the dominant staphyloxanthin bands in the MRSA, a cell component not associated with the presence of methicillin resistance. This is the first step toward the development of label-free Raman spectroscopy for the discrimination of MRSA and MSSA using single-cell analysis with 532 nm excitation. IMPORTANCE Label-free Raman spectra capture the high chemical complexity of bacterial cells. Many different Raman approaches have been developed using different excitation wavelength and cell analysis methods. This study highlights the major importance of selecting the most suitable Raman approach, capable of providing spectral features that can be associated with the cell mechanism under investigation. It is shown that the approach of choice for differentiating MRSA from MSSA should be single-cell analysis with 532 nm excitation since it captures the difference in the overall biochemical composition. These results should be taken into consideration in future studies aiming for the development of label-free Raman spectroscopy as a clinical analytical tool for antimicrobial resistance determination.
Keywords: 532-nm single-cell analysis; 785 nm fiber probe; MRSA; Raman microscopy; Raman spectroscopy; UV resonance Raman spectroscopy; single-cell analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Surface-enhanced Raman spectroscopy for comparison of biochemical profile of bacteriophage sensitive and resistant methicillin-resistant Staphylococcus aureus (MRSA) strains.Spectrochim Acta A Mol Biomol Spectrosc. 2024 Apr 5;310:123968. doi: 10.1016/j.saa.2024.123968. Epub 2024 Jan 30. Spectrochim Acta A Mol Biomol Spectrosc. 2024. PMID: 38330510
-
[Investigation of macrolide, lincosamide and streptogramin B resistance in Staphylococcus aureus strains isolated from clinical samples by phenotypical and genotypical methods].Mikrobiyol Bul. 2015 Jan;49(1):1-14. Mikrobiyol Bul. 2015. PMID: 25706726 Turkish.
-
Surface-enhanced Raman scattering method for the identification of methicillin-resistant Staphylococcus aureus using positively charged silver nanoparticles.Mikrochim Acta. 2019 Jan 12;186(2):102. doi: 10.1007/s00604-018-3150-6. Mikrochim Acta. 2019. PMID: 30637528
-
Antibiotic Resistance in the Treatment of Staphylococcus aureus Keratitis: a 20-Year Review.Cornea. 2015 Jun;34(6):698-703. doi: 10.1097/ICO.0000000000000431. Cornea. 2015. PMID: 25811722 Free PMC article. Review.
-
Correlation Between Biofilm Formation and Antibiotic Resistance in MRSA and MSSA Isolated from Clinical Samples in Iran: A Systematic Review and Meta-Analysis.Microb Drug Resist. 2020 Sep;26(9):1071-1080. doi: 10.1089/mdr.2020.0001. Epub 2020 Mar 10. Microb Drug Resist. 2020. PMID: 32159447
Cited by
-
A Pilot Study on Single-Cell Raman Spectroscopy Combined with Machine Learning for Phenotypic Characterization of Staphylococcus aureus.Microorganisms. 2025 Jun 8;13(6):1333. doi: 10.3390/microorganisms13061333. Microorganisms. 2025. PMID: 40572221 Free PMC article.
-
Recent advances in surface enhanced Raman spectroscopy for bacterial pathogen identifications.J Adv Res. 2023 Sep;51:91-107. doi: 10.1016/j.jare.2022.11.010. Epub 2022 Dec 19. J Adv Res. 2023. PMID: 36549439 Free PMC article.
-
Nanoplasmonic SERS on fidget spinner for digital bacterial identification.Microsyst Nanoeng. 2025 Mar 3;11(1):38. doi: 10.1038/s41378-025-00870-1. Microsyst Nanoeng. 2025. PMID: 40025021 Free PMC article.
-
Antibiotic Resistance Diagnosis in ESKAPE Pathogens-A Review on Proteomic Perspective.Diagnostics (Basel). 2023 Mar 7;13(6):1014. doi: 10.3390/diagnostics13061014. Diagnostics (Basel). 2023. PMID: 36980322 Free PMC article. Review.
-
Molecular Characteristics of Methicillin-Resistant and Susceptible Staphylococcus aureus from Pediatric Patients in Eastern China.Pathogens. 2023 Apr 2;12(4):549. doi: 10.3390/pathogens12040549. Pathogens. 2023. PMID: 37111435 Free PMC article.
References
-
- Kourtis AP, Hatfield K, Baggs J, Mu Y, See I, Epson E, Nadle J, Kainer MA, Dumyati G, Petit S, Ray SM, Ham D, Capers C, Ewing H, Coffin N, McDonald LC, Jernigan J, Cardo D, Emerging Infections Program MRSA author group . 2019. Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections-United States. MMWR Morb Mortal Wkly Rep 68:214–219. doi:10.15585/mmwr.mm6809e1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

