Climate-Endangered Arctic Epishelf Lake Harbors Viral Assemblages with Distinct Genetic Repertoires
- PMID: 36005820
- PMCID: PMC9469726
- DOI: 10.1128/aem.00228-22
Climate-Endangered Arctic Epishelf Lake Harbors Viral Assemblages with Distinct Genetic Repertoires
Abstract
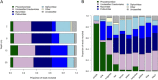
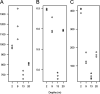
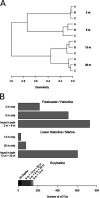
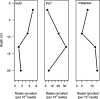
Milne Fiord, located on the coastal margin of the Last Ice Area (LIA) in the High Arctic (82°N, Canada), harbors an epishelf lake, a rare type of ice-dependent ecosystem in which a layer of freshwater overlies marine water connected to the open ocean. This microbe-dominated ecosystem faces catastrophic change due to the deterioration of its ice environment related to warming temperatures. We produced the first assessment of viral abundance, diversity, and distribution in this vulnerable ecosystem and explored the niches available for viral taxa and the functional genes underlying their distribution. We found that the viral community in the freshwater layer was distinct from, and more diverse than, the community in the underlying seawater and contained a different set of putative auxiliary metabolic genes, including the sulfur starvation-linked gene tauD and the gene coding for patatin-like phospholipase. The halocline community resembled the freshwater more than the marine community, but harbored viral taxa unique to this layer. We observed distinct viral assemblages immediately below the halocline, at a depth that was associated with a peak of prasinophyte algae and the viral family Phycodnaviridae. We also assembled 15 complete circular genomes, including a putative Pelagibacter phage with a marine distribution. It appears that despite its isolated and precarious situation, the varied niches in this epishelf lake support a diverse viral community, highlighting the importance of characterizing underexplored microbiota in the Last Ice Area before these ecosystems undergo irreversible change. IMPORTANCE Viruses are key to understanding polar aquatic ecosystems, which are dominated by microorganisms. However, studies of viral communities are challenging to interpret because the vast majority of viruses are known only from sequence fragments, and their taxonomy, hosts, and genetic repertoires are unknown. Our study establishes a basis for comparison that will advance understanding of viral ecology in diverse global environments, particularly in the High Arctic. Rising temperatures in this region mean that researchers have limited time remaining to understand the biodiversity and biogeochemical cycles of ice-dependent environments and the consequences of these rapid, irreversible changes. The case of the Milne Fiord epishelf lake has special urgency because of the rarity of this type of "floating lake" ecosystem and its location in the Last Ice Area, a region of thick sea ice with global importance for conservation efforts.
Keywords: Arctic; Caudovirales; Last Ice Area; Pelagibacter phage; Phycodnaviridae; auxiliary metabolic genes; epishelf lake; viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- IPCC. 2018. Global warming of 1.5°C. Special report. https://www.ipcc.ch/sr15/.
-
- Newton R, Pfirman S, Tremblay LB, DeRepentigny P. 2020. Defining the “ice shed” of the Arctic Ocean’s Last Ice Area and its future evolution. Earths Future 9:e2021EF001988.
-
- Vincent WF, Gibson JAE, Jeffries MO. 2001. Ice-shelf collapse, climate change, and habitat loss in the Canadian high Arctic. Polar Rec 37:133–142. 10.1017/S0032247400026954. - DOI
-
- Laybourn-Parry J, Wadham JL. 2014. Antarctic lakes. Oxford University Press, Oxford, United Kingdom.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

