The localization of PHRAGMOPLAST ORIENTING KINESIN1 at the division site depends on the microtubule-binding proteins TANGLED1 and AUXIN-INDUCED IN ROOT CULTURES9 in Arabidopsis
- PMID: 36005863
- PMCID: PMC9614452
- DOI: 10.1093/plcell/koac266
The localization of PHRAGMOPLAST ORIENTING KINESIN1 at the division site depends on the microtubule-binding proteins TANGLED1 and AUXIN-INDUCED IN ROOT CULTURES9 in Arabidopsis
Abstract
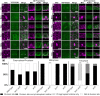
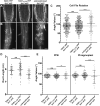
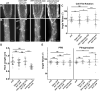
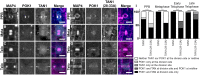
Proper plant growth and development require spatial coordination of cell divisions. Two unrelated microtubule-binding proteins, TANGLED1 (TAN1) and AUXIN-INDUCED IN ROOT CULTURES9 (AIR9), are together required for normal growth and division plane orientation in Arabidopsis (Arabidopsis thaliana). The tan1 air9 double mutant has synthetic growth and division plane orientation defects, while single mutants lack obvious defects. Here we show that the division site-localized protein, PHRAGMOPLAST ORIENTING KINESIN1 (POK1), was aberrantly lost from the division site during metaphase and telophase in the tan1 air9 mutant. Since TAN1 and POK1 interact via the first 132 amino acids of TAN1 (TAN11-132), we assessed the localization and function of TAN11-132 in the tan1 air9 double mutant. TAN11-132 rescued tan1 air9 mutant phenotypes and localized to the division site during telophase. However, replacing six amino-acid residues within TAN11-132, which disrupted the POK1-TAN1 interaction in the yeast-two-hybrid system, caused loss of both rescue and division site localization of TAN11-132 in the tan1 air9 mutant. Full-length TAN1 with the same alanine substitutions had defects in phragmoplast guidance and reduced TAN1 and POK1 localization at the division site but rescued most tan1 air9 mutant phenotypes. Together, these data suggest that TAN1 and AIR9 are required for POK1 localization, and yet unknown proteins may stabilize TAN1-POK1 interactions.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures



References
-
- Buschmann H, Chan J, Sanchez-Pulido L, Andrade-Navarro MA, Doonan JH, Lloyd CW (2006) Microtubule-associated AIR9 recognizes the cortical division site at preprophase and cell-plate insertion. Curr Biol 16: 1938–1943 - PubMed
-
- Buschmann H, Dols J, Kopischke S, Peña EJ, Andrade-Navarro MA, Heinlein M, Szymanski DB, Zachgo S, Doonan JH, Lloyd CW (2015) Arabidopsis KCBP interacts with AIR9 but stays in the cortical division zone throughout mitosis via its MyTH4-FERM domain. J Cell Sci 128: 2033–2046 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

