A reciprocal translocation involving Aspergillus nidulans snxAHrb1/Gbp2 and gyfA uncovers a new regulator of the G2-M transition and reveals a role in transcriptional repression for the setBSet2 histone H3-lysine-36 methyltransferase
- PMID: 36005881
- PMCID: PMC9526064
- DOI: 10.1093/genetics/iyac130
A reciprocal translocation involving Aspergillus nidulans snxAHrb1/Gbp2 and gyfA uncovers a new regulator of the G2-M transition and reveals a role in transcriptional repression for the setBSet2 histone H3-lysine-36 methyltransferase
Abstract
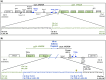
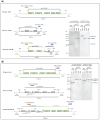
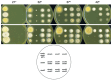
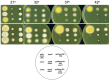
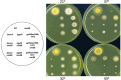
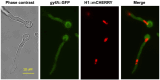
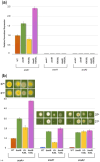
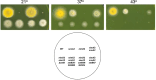
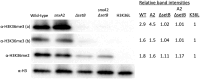
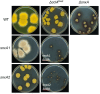
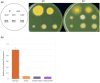
Aspergillus nidulans snxA, an ortholog of Saccharomyces cerevisiae Hrb1/Gbp2 messenger RNA shuttle proteins, is-in contrast to budding yeast-involved in cell cycle regulation, in which snxA1 and snxA2 mutations as well as a snxA deletion specifically suppress the heat sensitivity of mutations in regulators of the CDK1 mitotic induction pathway. snxA mutations are strongly cold sensitive, and at permissive temperature snxA mRNA and protein expression are strongly repressed. Initial attempts to identify the causative snxA mutations revealed no defects in the SNXA protein. Here, we show that snxA1/A2 mutations resulted from an identical chromosome I-II reciprocal translocation with breakpoints in the snxA first intron and the fourth exon of a GYF-domain gene, gyfA. Surprisingly, a gyfA deletion and a reconstructed gyfA translocation allele suppressed the heat sensitivity of CDK1 pathway mutants in a snxA+ background, demonstrating that 2 unrelated genes, snxA and gyfA, act through the CDK1-CyclinB axis to restrain the G2-M transition, and for the first time identifying a role in G2-M regulation for a GYF-domain protein. To better understand snxA1/A2-reduced expression, we generated suppressors of snxA cold sensitivity in 2 genes: (1) loss of the abundant nucleolar protein Nsr1/nucleolin bypassed the requirement for snxA and (2) loss of the Set2 histone H3 lysine36 (H3K36) methyltransferase or a nonmethylatable histone H3K36L mutant rescued hypomorphic snxA mutants by restoring full transcriptional proficiency, indicating that methylation of H3K36 acts normally to repress snxA transcription. These observations are in line with known Set2 functions in preventing excessive and cryptic transcription of active genes.
Keywords: H3K36me3; H3K4me3; cclABre2; gyfA; nsr1/nucleolin; nsrA; setB; snxAHrb1/Gbp2; GYF domain; translocation.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Azevedo C, Desfougères Y, Jiramongkol Y, Partington H, Trakansuebkul S, Singh J, Steck N, Jessen HK, Saiardi A. Development of a yeast model to study the contribution of vacuolar polyphosphate metabolism to lysine polyphosphorylation. J Biol Chem. 2020;295(6):1439–1451. 10.1074/jbc.RA119.011680 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

