Isolation, Characterization and Biological Action of Type-1 Ribosome-Inactivating Proteins from Tissues of Salsola soda L
- PMID: 36006228
- PMCID: PMC9412391
- DOI: 10.3390/toxins14080566
Isolation, Characterization and Biological Action of Type-1 Ribosome-Inactivating Proteins from Tissues of Salsola soda L
Abstract
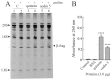
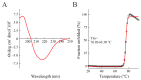
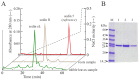
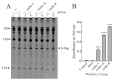
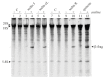
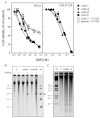
Ribosome-inactivating proteins (RIPs) are known as RNA N-glycosylases. They depurinate the major rRNA, damaging ribosomes and inhibiting protein synthesis. Here, new single-chain (type-1) RIPs named sodins were isolated from the seeds (five proteins), edible leaves (one protein) and roots (one protein) of Salsola soda L. Sodins are able to release Endo's fragment when incubated with rabbit and yeast ribosomes and inhibit protein synthesis in cell-free systems (IC50 = 4.83-79.31 pM). In addition, sodin 5, the major form isolated from seeds, as well as sodin eL and sodin R, isolated from edible leaves and roots, respectively, display polynucleotide:adenosine glycosylase activity and are cytotoxic towards the Hela and COLO 320 cell lines (IC50 = 0.41-1200 nM), inducing apoptosis. The further characterization of sodin 5 reveals that this enzyme shows a secondary structure similar to other type-1 RIPs and a higher melting temperature (Tm = 76.03 ± 0.30 °C) and is non-glycosylated, as other sodins are. Finally, we proved that sodin 5 possesses antifungal activity against Penicillium digitatum.
Keywords: agretti; antifungal activity; cytotoxicity; edible plants; protein purification; rRNA N-glycosylases.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

