A Long-Read Genome Assembly of a Native Mite in China Pyemotes zhonghuajia Yu, Zhang & He (Prostigmata: Pyemotidae) Reveals Gene Expansion in Toxin-Related Gene Families
- PMID: 36006233
- PMCID: PMC9415403
- DOI: 10.3390/toxins14080571
A Long-Read Genome Assembly of a Native Mite in China Pyemotes zhonghuajia Yu, Zhang & He (Prostigmata: Pyemotidae) Reveals Gene Expansion in Toxin-Related Gene Families
Abstract
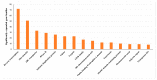
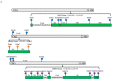
Pyemotes zhonghuajia Yu, Zhang & He (Prostigmata: Pyemotidae), discovered in China, has been demonstrated as a high-efficient natural enemy in controlling many agricultural and forestry pests. This mite injects toxins into the host (eggs, larvae, pupae, and adults), resulting in its paralyzation and then gets nourishment for reproductive development. These toxins have been approved to be mammal-safe, which have the potential to be used as biocontrol pesticides. Toxin proteins have been identified from many insects, especially those from the orders Scorpions and Araneae, some of which are now widely used as efficient biocontrol pesticides. However, toxin proteins in mites are not yet understood. In this study, we assembled the genome of P. zhonghuajia using PacBio technology and then identified toxin-related genes that are likely to be responsible for the paralytic process of P. zhonghuajia. The genome assembly has a size of 71.943 Mb, including 20 contigs with a N50 length of 21.248 Mb and a BUSCO completeness ratio of 90.6% (n = 1367). These contigs were subsequently assigned to three chromosomes. There were 11,183 protein coding genes annotated, which were assessed with 91.2% BUSCO completeness (n = 1066). Neurotoxin and dermonecrotic toxin gene families were significantly expanded within the genus of Pyemotes and they also formed several gene clusters on the chromosomes. Most of the genes from these two families and all of the three agatoxin genes were shown with higher expression in the one-day-old mites compared to the seven-day-pregnant mites, supporting that the one-day-old mites cause paralyzation and even death of the host. The identification of these toxin proteins may provide insights into how to improve the parasitism efficiency of this mite, and the purification of these proteins may be used to develop new biological pesticides.
Keywords: Pyemotes; genome annotation; protein; toxin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Predator-parasitoid interaction between Harmonia axyridis (Coleoptera: Coccinellidae) and Pyemotes zhonghuajia (Prostigmata: Pyemotidae) in aphid control.J Econ Entomol. 2025 Apr 26;118(2):505-513. doi: 10.1093/jee/toaf013. J Econ Entomol. 2025. PMID: 39985561
-
Optimizing cold storage of the ectoparasitic mite Pyemotes zhonghuajia (Acari: Pyemotidae), an efficient biological control agent of stem borers.Exp Appl Acarol. 2019 Jul;78(3):327-342. doi: 10.1007/s10493-019-00386-0. Epub 2019 Jun 27. Exp Appl Acarol. 2019. PMID: 31250238
-
The Biocontrol Agent Pyemotes zhonghuajia Has the Highest Lethal Weight Ratio Compared with Its Prey and the Most Dramatic Body Weight Change during Pregnancy.Insects. 2021 May 25;12(6):490. doi: 10.3390/insects12060490. Insects. 2021. PMID: 34070280 Free PMC article.
-
Pest management of postharvest potatoes: lethal, sublethal and transgenerational effects of the ectoparasitic mite Pyemotes zhonghuajia on the potato worm Phthorimaea operculella.Pest Manag Sci. 2023 Dec;79(12):5250-5259. doi: 10.1002/ps.7730. Epub 2023 Aug 31. Pest Manag Sci. 2023. PMID: 37595072
-
[Origin and evolution of parasitism in mites of the infraorder Eleutherengona (Acari: Prostigmata). Report I. Lower Raphignathae].Parazitologiia. 2008 Sep-Oct;42(5):337-59. Parazitologiia. 2008. PMID: 19065835 Review. Russian.
Cited by
-
Advanced Research on Animal Venoms in China.Toxins (Basel). 2023 Apr 6;15(4):272. doi: 10.3390/toxins15040272. Toxins (Basel). 2023. PMID: 37104210 Free PMC article.
References
-
- Yu L., Zhang Z.Q., He L. Two New Species of Pyemotes Closely Related to P. Tritici (Acari: Pyemotidae) Zootaxa. 2010;2723:1–40. doi: 10.11646/zootaxa.2723.1.1. - DOI
-
- He L.M., Jiao R., Xu C.X., Hao B.F., Han J.C., Yu L.C. Application of mtDNA COI Gene Sequence in Indentification of Pyemotes. J. Hebei Agric. Sci. 2010;14:46–50.
-
- Liu J.F., Tian T.A., Li X.L., Chen Y.C., Yu X.F., Tan X.F., Zhu Y., Yang M.F. Is Pyemotes Zhonghuajia (Acari: Pyemotidae) a Suitable Biological Control Agent against the Fall Armyworm Spodoptera Frugiperda (Lepidoptera: Noctuidae)? Syst. Appl. Acarol. 2020;25:649–657. doi: 10.11158/saa.25.4.5. - DOI
-
- Li L., He L., Yu L., He X.Z., Xu C., Jiao R., Zhang L., Liu J. Preliminary Study on the Potential of Pyemotes Zhonghuajia (Acari: Pyemotidae) in Biological Control of Aphis Citricola (Hemiptera: Aphididae) Syst. Appl. Acarol. 2019;24:1116–1120. doi: 10.11158/saa.24.6.12. - DOI
-
- De Sousa A.H., Mendonça G.R.Q., Lopes L.M., Faroni L.R.D. Widespread Infestation of Pyemotes Tritici (Acari: Pyemotidae) in Colonies of Seven Species of Stored-Product Insects. Genet. Mol. Res. 2020;19:1–5. doi: 10.4238/gmr18548. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

