Differences between Two Groups of Burmese Vipers (Viperidae: Azemiops) in the Proteomic Profiles, Immunoreactivity and Biochemical Functions of Their Venoms
- PMID: 36006235
- PMCID: PMC9416478
- DOI: 10.3390/toxins14080572
Differences between Two Groups of Burmese Vipers (Viperidae: Azemiops) in the Proteomic Profiles, Immunoreactivity and Biochemical Functions of Their Venoms
Abstract
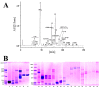
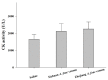
Two recently revised Azemiops snakes with apparent differences in their external appearances and skeletal morphologies but unclear genetic boundaries have been proposed. Some researchers have refrained from using the newly proposed taxonomy because these two "species" might be two clades corresponding to different geographical populations of Azemiops feae. To improve the understanding of the kinship of these two Burmese viper groups, more of their characteristics should be explored in depth. We performed a comparative analysis of the proteomic profiles and biochemical activities of snake venoms from these two groups (Sichuan A. feae and Zhejiang A. feae) and evaluated the immunorecognition capacity of commercial antivenoms toward them. Eight protein families were identified in venoms from these two groups, while phospholipase B was only detected in venom from Sichuan A. feae. These protein families displayed varying degrees of differences in relative abundance between venoms, and phospholipase A2 (Sichuan A. feae: 57.15%; Zhejiang A. feae: 65.94%) was the predominated component. Gloydius brevicaudus antivenom exhibited the strongest capacity to immunologically recognize these two venoms, but this was mainly limited to components with high molecular masses, some of which differed between venoms. Additionally, Zhejiang A. feae venom was more toxic than Sichuan A. feae venom, and the venoms expressed remarkable differences in enzymatic activities, probably resulting from the variation in the relative abundance of specific protein families. Our findings unveil differences between the two Burmese viper groups in terms of proteomic profiles, immunoreactivity, and the biochemical functions of their venoms. This information will facilitate the management of snakebites caused by these snakes.
Keywords: Azemiops; biochemical activity; proteome; taxonomy; venom.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Proteomic and biochemical analyses of short-tailed pit viper (Gloydius brevicaudus) venom: age-related variation and composition-activity correlation.J Proteomics. 2014 Jun 13;105:307-22. doi: 10.1016/j.jprot.2014.01.019. Epub 2014 Jan 31. J Proteomics. 2014. PMID: 24487038
-
Venomous snakes of Costa Rica: biological and medical implications of their venom proteomic profiles analyzed through the strategy of snake venomics.J Proteomics. 2014 Jun 13;105:323-39. doi: 10.1016/j.jprot.2014.02.020. Epub 2014 Feb 24. J Proteomics. 2014. PMID: 24576642 Review.
-
De Novo Genome Assembly Highlights the Role of Lineage-Specific Gene Duplications in the Evolution of Venom in Fea's Viper (Azemiops feae).Genome Biol Evol. 2022 Jul 2;14(7):evac082. doi: 10.1093/gbe/evac082. Genome Biol Evol. 2022. PMID: 35670514 Free PMC article.
-
Structures of Azemiops feae venom phospholipases and cys-rich-secretory protein and implications for taxonomy and toxinology.Toxicon. 2016 May;114:31-9. doi: 10.1016/j.toxicon.2016.02.014. Epub 2016 Feb 18. Toxicon. 2016. PMID: 26908291
-
Mass spectrometric analysis to unravel the venom proteome composition of Indian snakes: opening new avenues in clinical research.Expert Rev Proteomics. 2020 May;17(5):411-423. doi: 10.1080/14789450.2020.1778471. Epub 2020 Jun 24. Expert Rev Proteomics. 2020. PMID: 32579411 Review.
Cited by
-
First Look at the Venoms of Two Sinomicrurus Snakes: Differences in Yield, Proteomic Profiles, and Immunorecognition by Commercial Antivenoms.Toxins (Basel). 2025 Jan 2;17(1):19. doi: 10.3390/toxins17010019. Toxins (Basel). 2025. PMID: 39852972 Free PMC article.
-
Phylogeny-Related Variations in Venomics: A Test in a Subset of Habu Snakes (Protobothrops).Toxins (Basel). 2023 May 21;15(5):350. doi: 10.3390/toxins15050350. Toxins (Basel). 2023. PMID: 37235384 Free PMC article.
References
-
- Gutiérrez J.M., Calvete J.J., Habib A.G., Harrison R.A., Williams D.J., Warrell D.A. Snakebite envenoming. Nat. Rev. Dis. Primers. 2017;3:17063. - PubMed
-
- Fry B.G., Scheib H., van der Weerd L., Young B., McNaughtan J., Ramjan S.F.R., Vidal N., Poelmann R.E., Norman J.A. Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia) Mol. Cell. Proteom. 2008;7:215–246. doi: 10.1074/mcp.M700094-MCP200. - DOI - PubMed
-
- Campos P.F., Andrade-Silva D., Zelanis A., Paes Leme A.F., Rocha M.M., Menezes M.C., Serrano S.M., Junqueira-de-Azevedo Ide L. Trends in the evolution of snake toxins underscored by an integrative omics approach to profile the venom of the colubrid Phalotris mertensi. Genom. Biol. Evol. 2016;8:2266–2287. doi: 10.1093/gbe/evw149. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

