Trace the History of HIV and Predict Its Future through Genetic Sequences
- PMID: 36006282
- PMCID: PMC9416588
- DOI: 10.3390/tropicalmed7080190
Trace the History of HIV and Predict Its Future through Genetic Sequences
Abstract
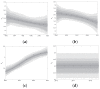
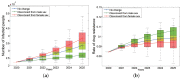
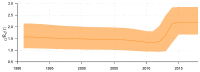
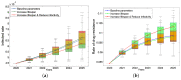
Traditional methods of quantifying epidemic spread are based on surveillance data. The most widely used surveillance data are normally incidence data from case reports and hospital records, which are normally susceptible to human error, and sometimes, they even can be seriously error-prone and incomplete when collected during a destructive epidemic. In this manuscript, we introduce a new method to study the spread of infectious disease. We gave an example of how to use this method to predict the virus spreading using the HIV gene sequences data of China. First, we applied Bayesian inference to gene sequences of two main subtypes of the HIV virus to infer the effective reproduction number (GRe(t)) to trace the history of HIV transmission. Second, a dynamic model was established to forecast the spread of HIV medication resistance in the future and also obtain its effective reproduction number (MRe(t)). Through fitting the two effective reproduction numbers obtained from the two separate ways above, some crucial parameters for the dynamic model were obtained. Simply raising the treatment rate has no impact on lowering the infection rate, according to the dynamics model research, but would instead increase the rate of medication resistance. The negative relationship between the prevalence of HIV and the survivorship of infected individuals following treatment may be to blame for this. Reducing the MSM population's number of sexual partners is a more efficient strategy to reduce transmission per the sensitivity analysis.
Keywords: Bayesian phylogenetic method; HIV/AIDS; dynamic model; effective reproductive number; primary drug resistance; transmitted drug resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Sun Z.Y., Wang K., Fan H. Progress in the development and control of drug resistance in highly active antiretroviral therapy (HAART) for HIV/AIDS. Jiangsu Prev. Med. 2019;30:159–162.
-
- Franois C., Allan J.H. HIV Drug Resistance. N. Engl. J. Med. 2004;350:1023–1035. - PubMed
-
- Ragonnet-Cronin M., Jackson C., Bradley-Stewart A., Aitken C., McAuley A., Palmateer N., Gunson R., Goldberg D., Milosevic C., Brown A.J.L. Recent and Rapid Transmission of HIV among People Who Inject Drugs in Scotland Revealed through Phylogenetic Analysis. J. Infect. Dis. 2018;217:1875–1882. doi: 10.1093/infdis/jiy130. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

