The phospho-landscape of the survival of motoneuron protein (SMN) protein: relevance for spinal muscular atrophy (SMA)
- PMID: 36006469
- PMCID: PMC11071818
- DOI: 10.1007/s00018-022-04522-9
The phospho-landscape of the survival of motoneuron protein (SMN) protein: relevance for spinal muscular atrophy (SMA)
Abstract
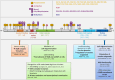
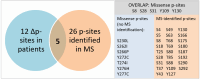
Spinal muscular atrophy (SMA) is caused by low levels of the survival of motoneuron (SMN) Protein leading to preferential degeneration of lower motoneurons in the ventral horn of the spinal cord and brain stem. However, the SMN protein is ubiquitously expressed and there is growing evidence of a multisystem phenotype in SMA. Since a loss of SMN function is critical, it is important to decipher the regulatory mechanisms of SMN function starting on the level of the SMN protein itself. Posttranslational modifications (PTMs) of proteins regulate multiple functions and processes, including activity, cellular trafficking, and stability. Several PTM sites have been identified within the SMN sequence. Here, we map the identified SMN PTMs highlighting phosphorylation as a key regulator affecting localization, stability and functions of SMN. Furthermore, we propose SMN phosphorylation as a crucial factor for intracellular interaction and cellular distribution of SMN. We outline the relevance of phosphorylation of the spinal muscular atrophy (SMA) gene product SMN with regard to basic housekeeping functions of SMN impaired in this neurodegenerative disease. Finally, we compare SMA patient mutations with putative and verified phosphorylation sites. Thus, we emphasize the importance of phosphorylation as a cellular modulator in a clinical perspective as a potential additional target for combinatorial SMA treatment strategies.
Keywords: Phosphorylation; Posttranslational modification (PTM); Spinal muscular atrophy (sMA); Survival of motoneuron (SMN) protein.
© 2022. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
All authors declared no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

