In vivo base editing by a single i.v. vector injection for treatment of hemoglobinopathies
- PMID: 36006707
- PMCID: PMC9675455
- DOI: 10.1172/jci.insight.162939
In vivo base editing by a single i.v. vector injection for treatment of hemoglobinopathies
Abstract
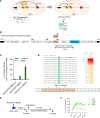
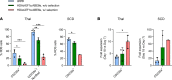
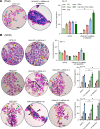
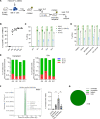
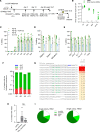
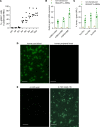
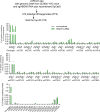
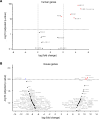
Individuals with β-thalassemia or sickle cell disease and hereditary persistence of fetal hemoglobin (HPFH) possessing 30% fetal hemoglobin (HbF) appear to be symptom free. Here, we used a nonintegrating HDAd5/35++ vector expressing a highly efficient and accurate version of an adenine base editor (ABE8e) to install, in vivo, a -113 A>G HPFH mutation in the γ-globin promoters in healthy CD46/β-YAC mice carrying the human β-globin locus. Our in vivo hematopoietic stem cell (HSC) editing/selection strategy involves only s.c. and i.v. injections and does not require myeloablation and HSC transplantation. In vivo HSC base editing in CD46/β-YAC mice resulted in > 60% -113 A>G conversion, with 30% γ-globin of β-globin expressed in 70% of erythrocytes. Importantly, no off-target editing at sites predicted by CIRCLE-Seq or in silico was detected. Furthermore, no critical alterations in the transcriptome of in vivo edited mice were found by RNA-Seq. In vitro, in HSCs from β-thalassemia and patients with sickle cell disease, transduction with the base editor vector mediated efficient -113 A>G conversion and reactivation of γ-globin expression with subsequent phenotypic correction of erythroid cells. Because our in vivo base editing strategy is safe and technically simple, it has the potential for clinical application in developing countries where hemoglobinopathies are prevalent.
Keywords: Gene therapy; Hematology; Hematopoietic stem cells; Monogenic diseases; Stem cells.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

