Constructive connectomics: How neuronal axons get from here to there using gene-expression maps derived from their family trees
- PMID: 36006873
- PMCID: PMC9409546
- DOI: 10.1371/journal.pcbi.1010382
Constructive connectomics: How neuronal axons get from here to there using gene-expression maps derived from their family trees
Abstract
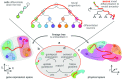
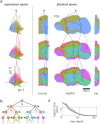
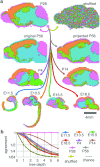
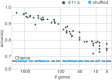
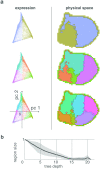
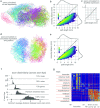
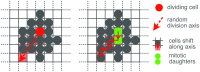
During brain development, billions of axons must navigate over multiple spatial scales to reach specific neuronal targets, and so build the processing circuits that generate the intelligent behavior of animals. However, the limited information capacity of the zygotic genome puts a strong constraint on how, and which, axonal routes can be encoded. We propose and validate a mechanism of development that can provide an efficient encoding of this global wiring task. The key principle, confirmed through simulation, is that basic constraints on mitoses of neural stem cells-that mitotic daughters have similar gene expression to their parent and do not stray far from one another-induce a global hierarchical map of nested regions, each marked by the expression profile of its common progenitor population. Thus, a traversal of the lineal hierarchy generates a systematic sequence of expression profiles that traces a staged route, which growth cones can follow to their remote targets. We have analyzed gene expression data of developing and adult mouse brains published by the Allen Institute for Brain Science, and found them consistent with our simulations: gene expression indeed partitions the brain into a global spatial hierarchy of nested contiguous regions that is stable at least from embryonic day 11.5 to postnatal day 56. We use this experimental data to demonstrate that our axonal guidance algorithm is able to robustly extend arbors over long distances to specific targets, and that these connections result in a qualitatively plausible connectome. We conclude that, paradoxically, cell division may be the key to uniting the neurons of the brain.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Maximum Entropy Principle Underlies Wiring Length Distribution in Brain Networks.Cereb Cortex. 2021 Aug 26;31(10):4628-4641. doi: 10.1093/cercor/bhab110. Cereb Cortex. 2021. PMID: 33999124
-
Wiring of the brain by a range of guidance cues.Prog Neurobiol. 2002 Dec;68(6):393-407. doi: 10.1016/s0301-0082(02)00129-6. Prog Neurobiol. 2002. PMID: 12576293 Review.
-
Uncovering Statistical Links Between Gene Expression and Structural Connectivity Patterns in the Mouse Brain.Neuroinformatics. 2021 Oct;19(4):649-667. doi: 10.1007/s12021-021-09511-0. Epub 2021 Mar 11. Neuroinformatics. 2021. PMID: 33704701 Free PMC article.
-
Plasticity of neuronal connections in developing brains of mammals.Neurosci Res. 1992 Dec;15(4):235-53. doi: 10.1016/0168-0102(92)90045-e. Neurosci Res. 1992. PMID: 1337578 Review.
-
Morphology of pioneer and follower growth cones in the developing cerebral cortex.J Neurobiol. 1991 Sep;22(6):629-42. doi: 10.1002/neu.480220608. J Neurobiol. 1991. PMID: 1919567
Cited by
-
Hierarchical communities in the larval Drosophila connectome: Links to cellular annotations and network topology.Proc Natl Acad Sci U S A. 2024 Sep 17;121(38):e2320177121. doi: 10.1073/pnas.2320177121. Epub 2024 Sep 13. Proc Natl Acad Sci U S A. 2024. PMID: 39269775 Free PMC article.
-
Generating brain-wide connectome using synthetic axonal morphologies.Nat Commun. 2025 Jul 18;16(1):6611. doi: 10.1038/s41467-025-62030-3. Nat Commun. 2025. PMID: 40676034 Free PMC article.
-
Computational Generation of Long-range Axonal Morphologies.Neuroinformatics. 2025 Jan 10;23(1):3. doi: 10.1007/s12021-024-09696-0. Neuroinformatics. 2025. PMID: 39792293 Free PMC article.
-
Estimating Brain Similarity Networks With Diffusion MRI.Hum Brain Mapp. 2025 Aug 1;46(11):e70313. doi: 10.1002/hbm.70313. Hum Brain Mapp. 2025. PMID: 40782044 Free PMC article.
-
A generative model of the connectome with dynamic axon growth.bioRxiv [Preprint]. 2024 Feb 28:2024.02.23.581824. doi: 10.1101/2024.02.23.581824. bioRxiv. 2024. Update in: Netw Neurosci. 2024 Dec 10;8(4):1192-1211. doi: 10.1162/netn_a_00397. PMID: 38464116 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

