Archetypal Analysis for population genetics
- PMID: 36007005
- PMCID: PMC9451066
- DOI: 10.1371/journal.pcbi.1010301
Archetypal Analysis for population genetics
Abstract
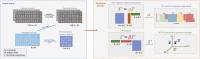
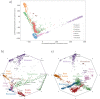
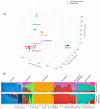
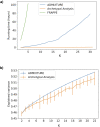
The estimation of genetic clusters using genomic data has application from genome-wide association studies (GWAS) to demographic history to polygenic risk scores (PRS) and is expected to play an important role in the analyses of increasingly diverse, large-scale cohorts. However, existing methods are computationally-intensive, prohibitively so in the case of nationwide biobanks. Here we explore Archetypal Analysis as an efficient, unsupervised approach for identifying genetic clusters and for associating individuals with them. Such unsupervised approaches help avoid conflating socially constructed ethnic labels with genetic clusters by eliminating the need for exogenous training labels. We show that Archetypal Analysis yields similar cluster structure to existing unsupervised methods such as ADMIXTURE and provides interpretative advantages. More importantly, we show that since Archetypal Analysis can be used with lower-dimensional representations of genetic data, significant reductions in computational time and memory requirements are possible. When Archetypal Analysis is run in such a fashion, it takes several orders of magnitude less compute time than the current standard, ADMIXTURE. Finally, we demonstrate uses ranging across datasets from humans to canids.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: CDB and AGI are co-founders of Galatea Bio Inc.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

