DMC1 attenuates RAD51-mediated recombination in Arabidopsis
- PMID: 36007010
- PMCID: PMC9451096
- DOI: 10.1371/journal.pgen.1010322
DMC1 attenuates RAD51-mediated recombination in Arabidopsis
Abstract
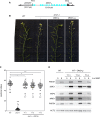
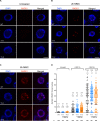
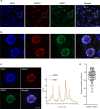
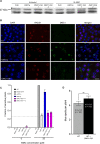
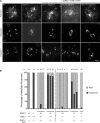
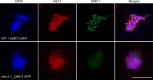
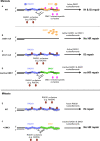
Ensuring balanced distribution of chromosomes in gametes, meiotic recombination is essential for fertility in most sexually reproducing organisms. The repair of the programmed DNA double strand breaks that initiate meiotic recombination requires two DNA strand-exchange proteins, RAD51 and DMC1, to search for and invade an intact DNA molecule on the homologous chromosome. DMC1 is meiosis-specific, while RAD51 is essential for both mitotic and meiotic homologous recombination. DMC1 is the main catalytically active strand-exchange protein during meiosis, while this activity of RAD51 is downregulated. RAD51 is however an essential cofactor in meiosis, supporting the function of DMC1. This work presents a study of the mechanism(s) involved in this and our results point to DMC1 being, at least, a major actor in the meiotic suppression of the RAD51 strand-exchange activity in plants. Ectopic expression of DMC1 in somatic cells renders plants hypersensitive to DNA damage and specifically impairs RAD51-dependent homologous recombination. DNA damage-induced RAD51 focus formation in somatic cells is not however suppressed by ectopic expression of DMC1. Interestingly, DMC1 also forms damage-induced foci in these cells and we further show that the ability of DMC1 to prevent RAD51-mediated recombination is associated with local assembly of DMC1 at DNA breaks. In support of our hypothesis, expression of a dominant negative DMC1 protein in meiosis impairs RAD51-mediated DSB repair. We propose that DMC1 acts to prevent RAD51-mediated recombination in Arabidopsis and that this down-regulation requires local assembly of DMC1 nucleofilaments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

