Molecular characterization of circulating Salmonella Typhi strains in an urban informal settlement in Kenya
- PMID: 36007074
- PMCID: PMC9451065
- DOI: 10.1371/journal.pntd.0010704
Molecular characterization of circulating Salmonella Typhi strains in an urban informal settlement in Kenya
Abstract
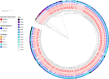
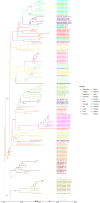
A high burden of Salmonella enterica subspecies enterica serovar Typhi (S. Typhi) bacteremia has been reported from urban informal settlements in sub-Saharan Africa, yet little is known about the introduction of these strains to the region. Understanding regional differences in the predominant strains of S. Typhi can provide insight into the genomic epidemiology. We genetically characterized 310 S. Typhi isolates from typhoid fever surveillance conducted over a 12-year period (2007-2019) in Kibera, an urban informal settlement in Nairobi, Kenya, to assess the circulating strains, their antimicrobial resistance attributes, and how they relate to global S. Typhi isolates. Whole genome multi-locus sequence typing (wgMLST) identified 4 clades, with up to 303 pairwise allelic differences. The identified genotypes correlated with wgMLST clades. The predominant clade contained 290 (93.5%) isolates with a median of 14 allele differences (range 0-52) and consisted entirely of genotypes 4.3.1.1 and 4.3.1.2. Resistance determinants were identified exclusively in the predominant clade. Determinants associated with resistance to aminoglycosides were observed in 245 isolates (79.0%), sulphonamide in 243 isolates (78.4%), trimethoprim in 247 isolates (79.7%), tetracycline in 224 isolates (72.3%), chloramphenicol in 247 isolates (79.6%), β-lactams in 239 isolates (77.1%) and quinolones in 62 isolates (20.0%). Multidrug resistance (MDR) determinants (defined as determinants conferring resistance to ampicillin, chloramphenicol and cotrimoxazole) were found in 235 (75.8%) isolates. The prevalence of MDR associated genes was similar throughout the study period (2007-2012: 203, 76.3% vs 2013-2019: 32, 72.7%; Fisher's Exact Test: P = 0.5478, while the proportion of isolates harboring quinolone resistance determinants increased (2007-2012: 42, 15.8% and 2013-2019: 20, 45.5%; Fisher's Exact Test: P<0.0001) following a decline in S. Typhi in Kibera. Some isolates (49, 15.8%) harbored both MDR and quinolone resistance determinants. There were no determinants associated with resistance to cephalosporins or azithromycin detected among the isolates sequenced in this study. Plasmid markers were only identified in the main clade including IncHI1A and IncHI1B(R27) in 226 (72.9%) isolates, and IncQ1 in 238 (76.8%) isolates. Molecular clock analysis of global typhoid isolates and isolates from Kibera suggests that genotype 4.3.1 has been introduced multiple times in Kibera. Several genomes from Kibera formed a clade with genomes from Kenya, Malawi, South Africa, and Tanzania. The most recent common ancestor (MRCA) for these isolates was from around 1997. Another isolate from Kibera grouped with several isolates from Uganda, sharing a common ancestor from around 2009. In summary, S. Typhi in Kibera belong to four wgMLST clades one of which is frequently associated with MDR genes and this poses a challenge in treatment and control.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

