Functions of block of proliferation 1 during anterior development in Xenopus laevis
- PMID: 36007075
- PMCID: PMC9409556
- DOI: 10.1371/journal.pone.0273507
Functions of block of proliferation 1 during anterior development in Xenopus laevis
Abstract
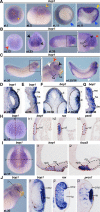
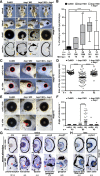
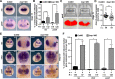
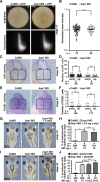
Block of proliferation 1 (Bop1) is a nucleolar protein known to be necessary for the assembly of the 60S subunit of ribosomes. Here, we show a specific bop1 expression in the developing anterior tissue of the South African clawed frog Xenopus laevis. Morpholino oligonucleotide-mediated knockdown approaches demonstrated that Bop1 is required for proper development of the cranial cartilage, brain, and the eyes. Furthermore, we show that bop1 knockdown leads to impaired retinal lamination with disorganized cell layers. Expression of neural crest-, brain-, and eye-specific marker genes was disturbed. Apoptotic and proliferative processes, which are known to be affected during ribosomal biogenesis defects, are not hindered upon bop1 knockdown. Because early Xenopus embryos contain a large store of maternal ribosomes, we considered if Bop1 might have a role independent of de novo ribosomal biogenesis. At early embryonic stages, pax6 expression was strongly reduced in bop1 morphants and synergy experiments indicate a common signaling pathway of the two molecules, Bop1 and Pax6. Our studies imply a novel function of Bop1 independent of ribosomal biogenesis.
Conflict of interest statement
NO authors have competing interests.
Figures



Similar articles
-
Bop1 is required to establish precursor domains of craniofacial tissues.Genesis. 2024 Feb;62(1):e23580. doi: 10.1002/dvg.23580. Epub 2023 Nov 16. Genesis. 2024. PMID: 37974491 Free PMC article.
-
Retinol binding protein 1 affects Xenopus anterior neural development via all-trans retinoic acid signaling.Dev Dyn. 2021 Aug;250(8):1096-1112. doi: 10.1002/dvdy.313. Epub 2021 Feb 24. Dev Dyn. 2021. PMID: 33570783
-
Peter Pan functions independently of its role in ribosome biogenesis during early eye and craniofacial cartilage development in Xenopus laevis.Development. 2011 Jun;138(11):2369-78. doi: 10.1242/dev.060160. Development. 2011. PMID: 21558383
-
Nosip functions during vertebrate eye and cranial cartilage development.Dev Dyn. 2018 Sep;247(9):1070-1082. doi: 10.1002/dvdy.24659. Epub 2018 Sep 9. Dev Dyn. 2018. PMID: 30055071
-
Using Xenopus to analyze neurocristopathies like Kabuki syndrome.Genesis. 2021 Feb;59(1-2):e23404. doi: 10.1002/dvg.23404. Epub 2020 Dec 22. Genesis. 2021. PMID: 33351273 Review.
Cited by
-
Bop1 is required to establish precursor domains of craniofacial tissues.Genesis. 2024 Feb;62(1):e23580. doi: 10.1002/dvg.23580. Epub 2023 Nov 16. Genesis. 2024. PMID: 37974491 Free PMC article.
-
Human pre-60S assembly factors link rRNA transcription to pre-rRNA processing.RNA. 2022 Nov 2;29(1):82-96. doi: 10.1261/rna.079149.122. Online ahead of print. RNA. 2022. PMID: 36323459 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

