Circular RNA hsa_circ_0007367 promotes the progression of pancreatic ductal adenocarcinoma by sponging miR-6820-3p and upregulating YAP1 expression
- PMID: 36008392
- PMCID: PMC9411600
- DOI: 10.1038/s41419-022-05188-8
Circular RNA hsa_circ_0007367 promotes the progression of pancreatic ductal adenocarcinoma by sponging miR-6820-3p and upregulating YAP1 expression
Abstract
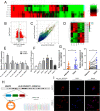
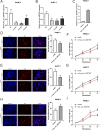
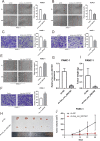
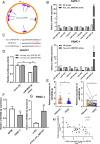
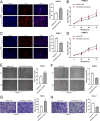
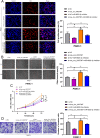
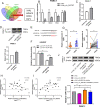
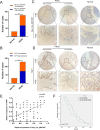
Circular RNAs (circRNAs) play critical regulatory roles in cancer biological processes. Nevertheless, the contributions and underlying mechanisms of circRNAs to pancreatic ductal adenocarcinoma (PDAC) remain largely unexplored. Dysregulated circRNAs between cancerous tissues and matched adjacent normal tissues were identified by circRNA microarray in PDAC. The biological effect of hsa_circ_007367 both in vitro and in vivo was demonstrated by gain- and loss-of-function experiments. Further, dual-luciferase reporter and RNA pull-down assays were performed to confirm the interaction among hsa_circ_007367, miR-6820-3p, and Yes-associated protein 1 (YAP1). The expression of hsa_circ_007367 and YAP1 were detected by in situ hybridization (ISH) and immunohistochemistry (IHC) using tissue microarray (TMA) in 128 PDAC samples. We first identified that a novel circRNA, hsa_circ_0007367, was markedly upregulated in PDAC tissues and cells. Functionally, in vivo and in vitro data indicated that hsa_circ_0007367 promotes the proliferation and metastasis of PDAC. Mechanistically, we confirmed that hsa_circ_0007367 could facilitate the expression of YAP1, a well-known oncogene, by sponging miR-6820-3p, which function as a tumor suppresser in PDAC cells. The results of ISH and IHC demonstrated that hsa_circ_0007367 and YAP1 were upregulated in PDAC tissues. Furthermore, clinical data showed that higher hsa_circ_0007367 expression was correlated with advanced histological grade and lymph node metastasis in PDAC patients. In conclusion, our findings reveal that hsa_circ_0007367 acts as an oncogene via modulating miR-6820-3p/YAP1 axis to promote the progression of PDAC, and suggest that hsa_circ_0007367 may serve as a potential therapeutic target for treatment of PDAC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b.J Cell Biochem. 2019 Mar;120(3):3780-3789. doi: 10.1002/jcb.27658. Epub 2018 Nov 1. J Cell Biochem. 2019. PMID: 30382592
-
Circ-0005105 activates COL11A1 by targeting miR-20a-3p to promote pancreatic ductal adenocarcinoma progression.Cell Death Dis. 2021 Jun 28;12(7):656. doi: 10.1038/s41419-021-03938-8. Cell Death Dis. 2021. PMID: 34183642 Free PMC article.
-
Upregulated circRNA hsa_circ_0071036 promotes tumourigenesis of pancreatic cancer by sponging miR-489 and predicts unfavorable characteristics and prognosis.Cell Cycle. 2021 Feb;20(4):369-382. doi: 10.1080/15384101.2021.1874684. Epub 2021 Jan 28. Cell Cycle. 2021. PMID: 33507122 Free PMC article.
-
Circular RNA_0000285: A novel double-edged sword circular RNA in human malignancies.Pathol Res Pract. 2023 Nov;251:154900. doi: 10.1016/j.prp.2023.154900. Epub 2023 Oct 20. Pathol Res Pract. 2023. PMID: 37871444 Review.
-
circWHSC1: A circular RNA piece in the human cancer puzzle.Pathol Res Pract. 2023 Sep;249:154730. doi: 10.1016/j.prp.2023.154730. Epub 2023 Jul 29. Pathol Res Pract. 2023. PMID: 37549517 Review.
Cited by
-
Down-regulation of long noncoding RNA HULC inhibits the inflammatory response in ankylosing spondylitis by reducing miR-556-5p-mediated YAP1 expression.J Orthop Surg Res. 2023 Jul 31;18(1):551. doi: 10.1186/s13018-023-04003-0. J Orthop Surg Res. 2023. PMID: 37525215 Free PMC article.
-
Circ_0003945: an emerging biomarker and therapeutic target for human diseases.Front Oncol. 2024 Apr 22;14:1275009. doi: 10.3389/fonc.2024.1275009. eCollection 2024. Front Oncol. 2024. PMID: 38711855 Free PMC article. Review.
-
CircATP13A1 (hsa_circ_0000919) promotes cell proliferation and metastasis and inhibits cell apoptosis in pancreatic ductal adenocarcinoma via the miR-186/miR-326/HMGA2 axis: implications for novel therapeutic targets.Am J Cancer Res. 2023 Nov 15;13(11):5610-5625. eCollection 2023. Am J Cancer Res. 2023. PMID: 38058810 Free PMC article.
-
Circular RNA in Pancreatic Cancer: Biogenesis, Mechanism, Function and Clinical Application.Int J Med Sci. 2025 Feb 28;22(7):1612-1629. doi: 10.7150/ijms.107773. eCollection 2025. Int J Med Sci. 2025. PMID: 40093798 Free PMC article. Review.
-
CircRNAs in Pancreatic Cancer: New Tools for Target Identification and Therapeutic Intervention.Cancer Genomics Proteomics. 2024 Jul-Aug;21(4):327-349. doi: 10.21873/cgp.20451. Cancer Genomics Proteomics. 2024. PMID: 38944427 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

