Extended longevity of DNA preservation in Levantine Paleolithic sediments, Sefunim Cave, Israel
- PMID: 36008437
- PMCID: PMC9411205
- DOI: 10.1038/s41598-022-17399-2
Extended longevity of DNA preservation in Levantine Paleolithic sediments, Sefunim Cave, Israel
Abstract
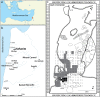
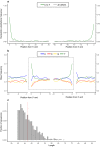
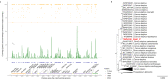
Paleogenomic research can elucidate the evolutionary history of human and faunal populations. Although the Levant is a key land-bridge between Africa and Eurasia, thus far, relatively little ancient DNA data has been generated from this region, since DNA degrades faster in warm climates. As sediments can be a source of ancient DNA, we analyzed 33 sediment samples from different sedimentological contexts in the Paleolithic layers of Sefunim Cave (Israel). Four contained traces of ancient Cervidae and Hyaenidae mitochondrial DNA. Dating by optical luminescence and radiocarbon indicates that the DNA comes from layers between 30,000 and 70,000 years old, surpassing theoretical expectations regarding the longevity of DNA deposited in such a warm environment. Both identified taxa are present in the zooarchaeological record of the site but have since gone extinct from the region, and a geoarchaeological study suggests little movement of the sediments after their deposition, lending further support to our findings. We provide details on the local conditions in the cave, which we hypothesize were particularly conducive to the long-term preservation of DNA-information that will be pertinent for future endeavors aimed at recovering ancient DNA from the Levant and other similarly challenging contexts.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Tchernov, E. The faunal sequence of the southwest Asian Middle Paleolithic in relation to hominid dispersal events. in Neandertals and Modern Humans in Western Asia 77–94 (Springer, 2002).
-
- Martínez-Navarro, B. Hippos, pigs, bovids, saber-toothed tigers, monkeys, and hominids: Dispersals through the Levantine corridor during late Pliocene and early Pleistocene times. in Human Paleoecology in the Levantine Corridor 37–52 (Oxbow Books, 2004).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

