Structural bioinformatics analysis of SARS-CoV-2 variants reveals higher hACE2 receptor binding affinity for Omicron B.1.1.529 spike RBD compared to wild type reference
- PMID: 36008461
- PMCID: PMC9406262
- DOI: 10.1038/s41598-022-18507-y
Structural bioinformatics analysis of SARS-CoV-2 variants reveals higher hACE2 receptor binding affinity for Omicron B.1.1.529 spike RBD compared to wild type reference
Abstract
To date, more than 263 million people have been infected with SARS-CoV-2 during the COVID-19 pandemic. In many countries, the global spread occurred in multiple pandemic waves characterized by the emergence of new SARS-CoV-2 variants. Here we report a sequence and structural-bioinformatics analysis to estimate the effects of amino acid substitutions on the affinity of the SARS-CoV-2 spike receptor binding domain (RBD) to the human receptor hACE2. This is done through qualitative electrostatics and hydrophobicity analysis as well as molecular dynamics simulations used to develop a high-precision empirical scoring function (ESF) closely related to the linear interaction energy method and calibrated on a large set of experimental binding energies. For the latest variant of concern (VOC), B.1.1.529 Omicron, our Halo difference point cloud studies reveal the largest impact on the RBD binding interface compared to all other VOC. Moreover, according to our ESF model, Omicron achieves a much higher ACE2 binding affinity than the wild type and, in particular, the highest among all VOCs except Alpha and thus requires special attention and monitoring.
© 2022. The Author(s).
Conflict of interest statement
V.D., K.K., A.S., An.K., D.N., Al.K., L.P., C.K., V.R., U.K. report working for Innophore. L.C., M.K., R.B., report working for Amazon Web Services, a company that also provides cloud computing services. K.G., G.S., C.C.G. report being shareholders of Innophore, an enzyme and drug discovery company. Additionally, G.S. and C.C.G. report being managing directors of Innophore. The research described here is scientifically and financially independent of the efforts in any of the above mentioned companies and open-science.
Figures
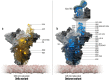


References
-
- WHO. Coronavirus disease (COVID-19) pandemic, https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (2021).
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

