A high-throughput drug screen reveals means to differentiate triple-negative breast cancer
- PMID: 36008466
- PMCID: PMC9507968
- DOI: 10.1038/s41388-022-02429-0
A high-throughput drug screen reveals means to differentiate triple-negative breast cancer
Abstract
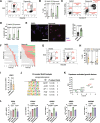
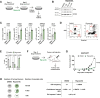
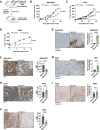
Plasticity delineates cancer subtypes with more or less favourable outcomes. In breast cancer, the subtype triple-negative lacks expression of major differentiation markers, e.g., estrogen receptor α (ERα), and its high cellular plasticity results in greater aggressiveness and poorer prognosis than other subtypes. Whether plasticity itself represents a potential vulnerability of cancer cells is not clear. However, we show here that cancer cell plasticity can be exploited to differentiate triple-negative breast cancer (TNBC). Using a high-throughput imaging-based reporter drug screen with 9 501 compounds, we have identified three polo-like kinase 1 (PLK1) inhibitors as major inducers of ERα protein expression and downstream activity in TNBC cells. PLK1 inhibition upregulates a cell differentiation program characterized by increased DNA damage, mitotic arrest, and ultimately cell death. Furthermore, cells surviving PLK1 inhibition have decreased tumorigenic potential, and targeting PLK1 in already established tumours reduces tumour growth both in cell line- and patient-derived xenograft models. In addition, the upregulation of genes upon PLK1 inhibition correlates with their expression in normal breast tissue and with better overall survival in breast cancer patients. Our results indicate that differentiation therapy based on PLK1 inhibition is a potential alternative strategy to treat TNBC.
© 2022. The Author(s).
Conflict of interest statement
MV, CJ, ALC, PAdM, TBP, MD, ASc, M-MC, KV, LS, RO, SDS, RM, declare no competing financial interests. AB, ASe. and MMSO are employees of F. Hoffmann–La Roche. JPC, VU, OG, IC, SL-R, GG, and DB are employees of Novartis. TR is an employee of Basilea. SM is part of a digital advisory board for Roche. WPW received research support from Takeda Pharmaceuticals International paid to the Swiss Group for Clinical Cancer Research (SAKK) and personal honoraria from Genomic Health. Support for meetings was paid to his institution from Sandoz, Genomic Health, Medtronic, Novartis Oncology, Pfizer and Eli Lilly. MB-A owns equities in and has received laboratory support and compensation from Novartis, and serves as a consultant for Basilea.
Figures


References
-
- Varga J, Greten FR. Cell plasticity in epithelial homeostasis and tumorigenesis. Nat Cell Biol. 2017;19:1133–41. - PubMed
-
- Clevers H, Watt FM. Annual Review of Biochemistry Defining Adult Stem Cells by Function, not by Phenotype. Annu Rev Biochem. 2018;87:1015–27.. - PubMed
-
- Koren S, Reavie L, Couto JP, de Silva D, Stadler MB, Roloff T, et al. PIK3CA H1047R induces multipotency and multi-lineage mammary tumours. Nature. 2015;525:114–8. - PubMed
-
- van Keymeulen A, Lee MY, Ousset M, Brohée S, Rorive S, Giraddi RR, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

