Expansion of the RNAStructuromeDB to include secondary structural data spanning the human protein-coding transcriptome
- PMID: 36008510
- PMCID: PMC9403969
- DOI: 10.1038/s41598-022-18699-3
Expansion of the RNAStructuromeDB to include secondary structural data spanning the human protein-coding transcriptome
Abstract
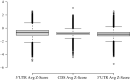
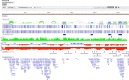
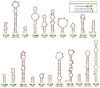
RNA plays vital functional roles in almost every component of biology, and these functional roles are often influenced by its folding into secondary and tertiary structures. An important role of RNA secondary structure is in maintaining proper gene regulation; therefore, making accurate predictions of the structures involved in these processes is important. In this study, we have expanded on our previous work that led to the creation of the RNAStructuromeDB. Unlike this previous study that analyzed the human genome at low resolution, we have now scanned the protein-coding human transcriptome at high (single nt) resolution. This provides more robust structure predictions for over 100,000 isoforms of known protein-coding genes. Notably, we also utilize the motif identification tool, ScanFold, to model structures with high propensity for ordered/evolved stability. All data have been uploaded to the RNAStructuromeDB, allowing for easy searching of transcripts, visualization of data tracks (via the Integrative Genomics Viewer or IGV), and download of ScanFold data-including unique highly-ordered motifs. Herein, we provide an example analysis of MAT2A to demonstrate the utility of ScanFold at finding known and novel secondary structures, highlighting regions of potential functionality, and guiding generation of functional hypotheses through use of the data.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

