Human alveolar progenitors generate dual lineage bronchioalveolar organoids
- PMID: 36008580
- PMCID: PMC9409623
- DOI: 10.1038/s42003-022-03828-5
Human alveolar progenitors generate dual lineage bronchioalveolar organoids
Abstract
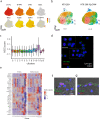
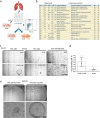
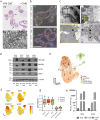
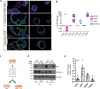
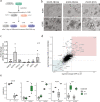
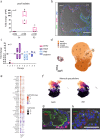
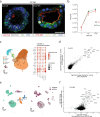
Mechanisms of epithelial renewal in the alveolar compartment remain incompletely understood. To this end, we aimed to characterize alveolar progenitors. Single-cell RNA-sequencing (scRNA-seq) analysis of the HTII-280+/EpCAM+ population from adult human lung revealed subclusters enriched for adult stem cell signature (ASCS) genes. We found that alveolar progenitors in organoid culture in vitro show phenotypic lineage plasticity as they can yield alveolar or bronchial cell-type progeny. The direction of the differentiation is dependent on the presence of the GSK-3β inhibitor, CHIR99021. By RNA-seq profiling of GSK-3β knockdown organoids we identified additional candidate target genes of the inhibitor, among others FOXM1 and EGF. This gives evidence of Wnt pathway independent regulatory mechanisms of alveolar specification. Following influenza A virus (IAV) infection organoids showed a similar response as lung tissue explants which confirms their suitability for studies of sequelae of pathogen-host interaction.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

