Wildfire-dependent changes in soil microbiome diversity and function
- PMID: 36008619
- PMCID: PMC9418001
- DOI: 10.1038/s41564-022-01203-y
Wildfire-dependent changes in soil microbiome diversity and function
Abstract
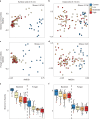
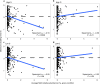
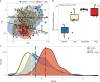
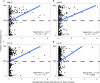
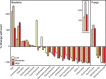
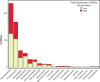
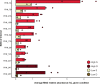
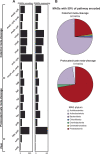
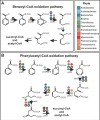
Forest soil microbiomes have crucial roles in carbon storage, biogeochemical cycling and rhizosphere processes. Wildfire season length, and the frequency and size of severe fires have increased owing to climate change. Fires affect ecosystem recovery and modify soil microbiomes and microbially mediated biogeochemical processes. To study wildfire-dependent changes in soil microbiomes, we characterized functional shifts in the soil microbiota (bacteria, fungi and viruses) across burn severity gradients (low, moderate and high severity) 1 yr post fire in coniferous forests in Colorado and Wyoming, USA. We found severity-dependent increases of Actinobacteria encoding genes for heat resistance, fast growth, and pyrogenic carbon utilization that might enhance post-fire survival. We report that increased burn severity led to the loss of ectomycorrhizal fungi and less tolerant microbial taxa. Viruses remained active in post-fire soils and probably influenced carbon cycling and biogeochemistry via turnover of biomass and ecosystem-relevant auxiliary metabolic genes. Our genome-resolved analyses link post-fire soil microbial taxonomy to functions and reveal the complexity of post-fire soil microbiome activity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Soil microbiota takes the heat.Nat Rev Microbiol. 2022 Nov;20(11):638. doi: 10.1038/s41579-022-00806-w. Nat Rev Microbiol. 2022. PMID: 36138155 No abstract available.
References
-
- Dennison PE, Brewer SC, Arnold JD, Moritz MA. Large wildfire trends in the western United States, 1984–2011. Geophys. Res. Lett. 2014;41:2928–2933.
-
- Higuera, P. E. & Abatzoglou, J. T. Record‐setting climate enabled the extraordinary 2020 fire season in the western United States. Glob. Change Biol. 10.1111/gcb.15388 (2020). - PubMed
-
- Parks, S. A. & Abatzoglou, J. T. Warmer and drier fire seasons contribute to increases in area burned at high severity in western US forests from 1985 to 2017. Geophys. Res. Lett.47, e2020GL089858 (2020).
-
- Benavides-Solorio JDD, MacDonald LH. Measurement and prediction of post-fire erosion at the hillslope scale, Colorado Front Range. Int. J. Wildl. Fire. 2005;14:457–474.
-
- Pierson DN, Robichaud PR, Rhoades CC, Brown RE. Soil carbon and nitrogen eroded after severe wildfire and erosion mitigation treatments. Int. J. Wildl. Fire. 2019;28:814–821.

